rebar
Dataset
A minimal example dataset is provided called toy1, which can be downloaded with:
rebar dataset download --name toy1 --tag custom --output-dir dataset/toy1
Mandatory
A rebar dataset consists of two mandatory parts:
-
reference.fasta: The reference genome.>Reference AAAAAAAAAAAAAAAAAAAA -
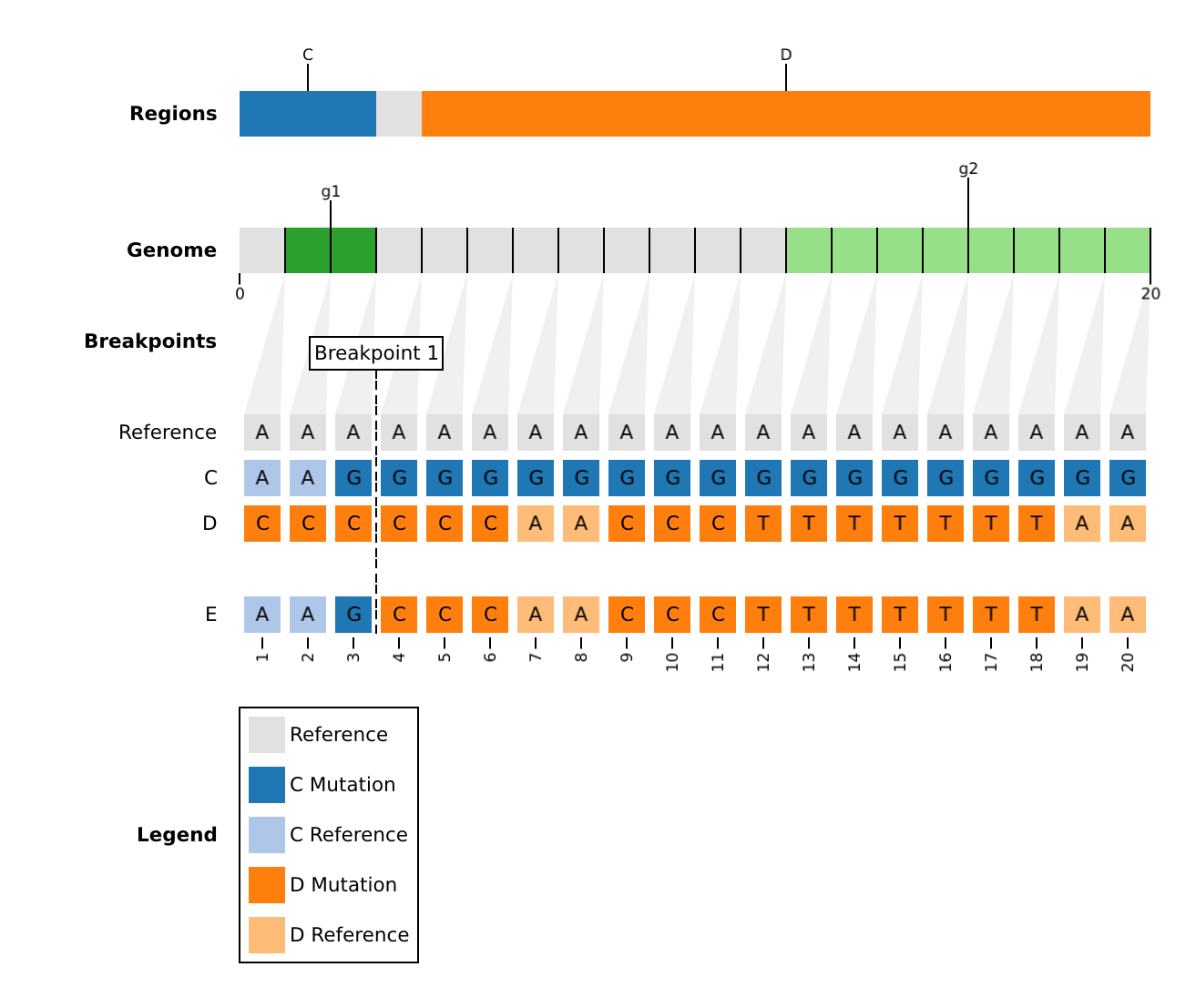
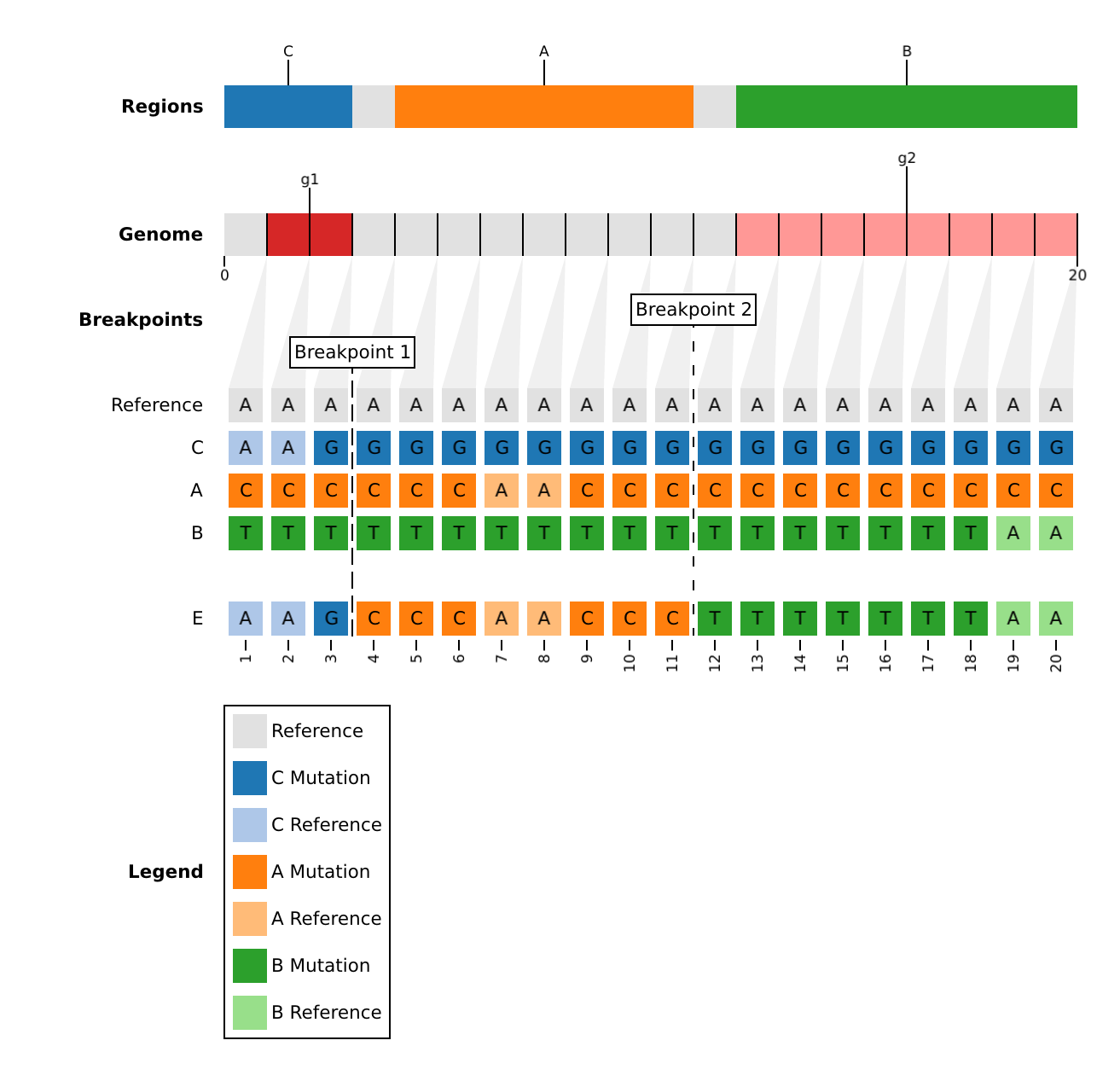
populations.fasta: Known populations (ex. clades, lineages) aligned to the reference.>A CCCCCCAACCCCCCCCCCCC >B TTTTTTTTTTTTTTTTTTAA >C AAGGGGGGGGGGGGGGGGGG >D CCCCCCAACCCTTTTTTTAA >E AAGCCCAACCCTTTTTTTAA
Optional
The following are optional components:
-
annotations.tsv: A table of genome annotations to add to the plot.gene abbreviation start end Gene1 g1 1 3 Gene2 g2 12 20 -
phylogeny.json: A phylogenetic graph which provides prior information about the evolutionary history. This is particularly useful if populations inpopulations.fastaare internal nodes or known recombinants.For example, an evolutionary history, in which population
Dis a recombinant, andEis a recursive recombinant (it has a parent that is also a recombinant).graph LR; root-->A; root-->B; root-->C; A-->D; B-->D; C-->E; D-->E; style D fill:#00758f style E fill:#00758fIs represented by the following phylogenetic graph:
{ "graph": { "nodes": ["root", "A", "B", "C", "D", "E" ], "edge_property": "directed", "edges": [ [ 0, 1, 1 ], [ 0, 2, 1 ], [ 0, 3, 1 ], [ 1, 4, 1 ], [ 2, 4, 1 ], [ 4, 5, 1 ], [ 3, 5, 1 ] ] } }Where
nodesare the list of node names in the tree (internal and external), andedgesare the branches between nodes. For example, the edge[0, 1, 1]connects node index 0 (“root”) to node index 1 (“A”) with a branch length of 1. Please note that branch lengths are not currently used inrebar'salgorithm. -
edge_cases.json: A list ofrebararguments to apply only to a particular population.In the dataset,
Eis a recursive recombinant between populationCand recombinantD. However, we could instead force it to be a recombinant betweenA,B, andCwith the following parameters:[ { "population": "E", "parents": [ "A", "B", "C"], "knockout": null, "mask": [0, 0], "max_iter": 3, "max_parents": 3, "min_parents": 3, "min_consecutive": 3, "min_length": 3, "min_subs": 1, "naive": false } ]Default Edge Case