rebar
Examples
Download a SARS-CoV-2 dataset, version controlled to the date 2023-11-30.
rebar dataset download \
--name sars-cov-2 \
--tag 2023-11-30 \
--output-dir dataset/sars-cov-2/2023-11-30
Populations
Use the names of dataset populations (ex. SARS-CoV-2 lineages) as input.
rebar run \
--dataset-dir dataset/sars-cov-2/2023-11-30 \
--populations "AY.4.2*,BA.5.2,XBC.1.6*,XBB.1.5.1,XBL" \
--output-dir output/example/population
rebar plot \
--run-dir output/example/population \
--annotations dataset/sars-cov-2/2023-11-30/annotations.tsv
The populations (--populations) can include any sequence name found in the dataset’s populations.fasta. For sars-cov-2, sequence names are the designated lineages.
The wildcard character (“*”) will include the population and all its descendants. The pattern (“X*”) will include only recombinants and their descendants. NOTE: If using “*”, make sure to use quotes (ex. --lineages "XBC*,XBB.1.16*")!
Alignment
Use an alignment of genomes as input.
wget https://raw.githubusercontent.com/phac-nml/rebar/main/data/example2.fasta
rebar run \
--dataset-dir dataset/sars-cov-2/2023-11-30 \
--alignment example2.fasta \
--output-dir output/example/alignment
rebar plot \
--run-dir output/example/alignment \
--annotations dataset/sars-cov-2/2023-11-30/annotations.tsv
Please note that the --alignment should be aligned to the same reference as in the dataset reference.fasta! We strongly recommend nextclade.
Debug
To understand the inner-workings of the rebar algorithm, you can enable the debugging log with --verbosity debug. This is an INCREDIBLY verbose log on the dataset searches and hypothesis testing. We recommend only using this for a small number of input parents/populations.
rebar run \
--dataset-dir dataset/sars-cov-2/2023-11-30 \
--populations "XD" \
--output-dir output/example/parents \
--verbosity debug
Parents
By default, rebar will consider all populations in the dataset as possible parents. If you would like to see the evidence for specific parents, you can restrict the parent search with --parents. For example, the recombinant XD is designated as having parents BA.1 (Omicron) and B.1.617.2 (generic Delta). But you might be interested in forcing it to evaluate a more specific Delta parent (ex. AY.4).
rebar run \
--dataset-dir dataset/sars-cov-2/2023-11-30 \
--populations "XD" \
--parents "AY.4*,BA.1" \
--output-dir output/example/parents \
--verbosity debug
rebar plot \
--run-dir output/example/parents \
--annotations dataset/sars-cov-2/2023-11-30/annotations.tsv
Knockout
Conversely to selecting specific parents, you can perform a ‘knockout’ experiment to remove populations from the dataset. For example, we might be interested in what the SARS-CoV-2 recombinant XBB would have been classified as before it became a designated lineage.
rebar run \
--dataset-dir dataset/sars-cov-2/2023-11-30 \
--populations "XBB" \
--knockout "XBB*" \
--output-dir output/example/knockout
rebar plot \
--run-dir output/example/knockout \
--annotations dataset/sars-cov-2/2023-11-30/annotations.tsv
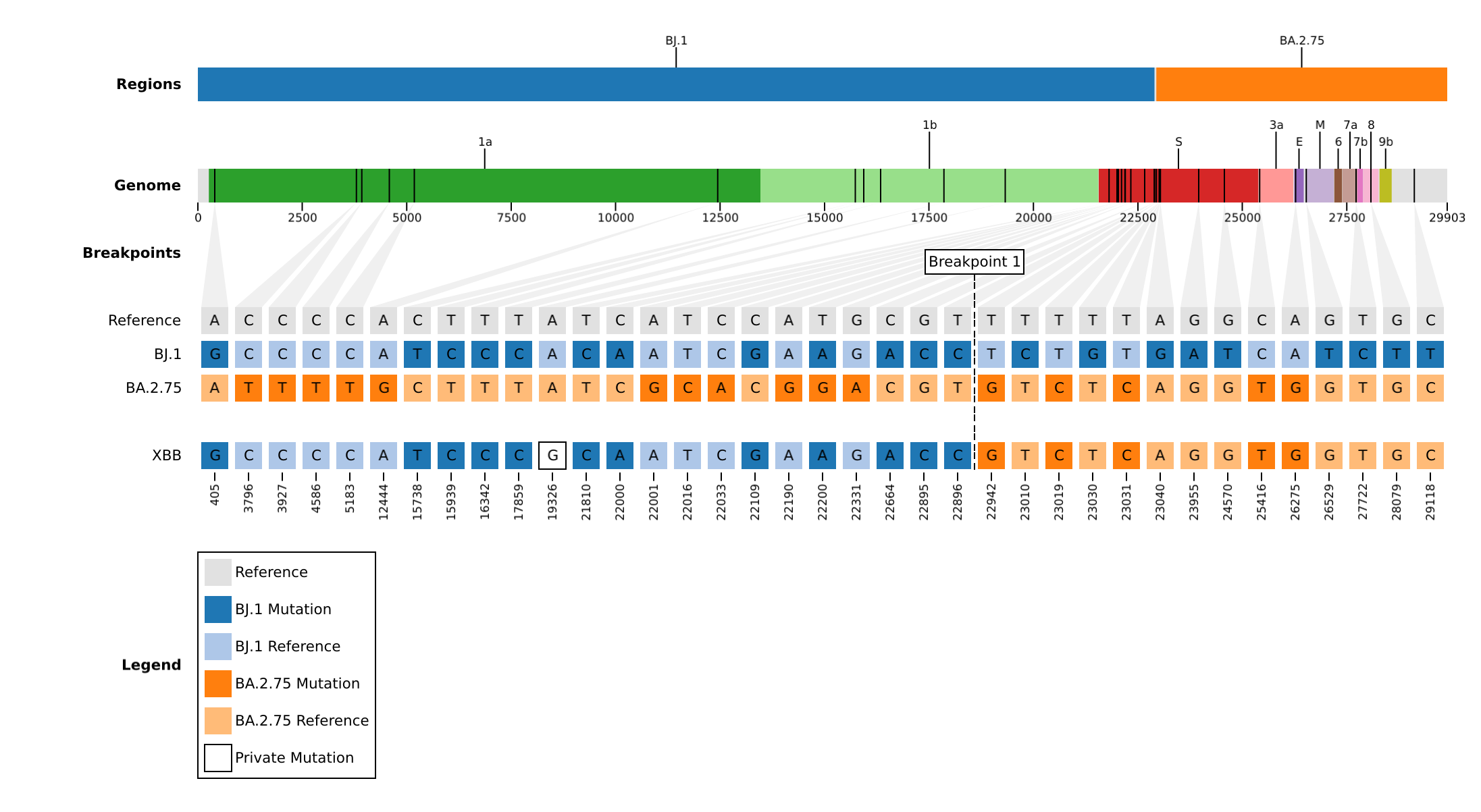
The linelist (output/example/knockout/linelist.tsv) reveals that:
XBBis descendant ofBJ.1(BA.2.10.1.1), specifically a novel recombinant betweenBJ.1andBA.2.75.-
The
substitutionscolumn reveals:- 5 substitutions came from the
BA.2.75parent:T22942G,T23019C,T23031C,C25416T,A26275G. - 1 substitution came from neither parent (private):
A19326G
- 5 substitutions came from the
- This can be used to contribute evidence for a new lineage proposal in the pango-designation respository.
| strain | validate | validate_details | population | recombinant | parents | breakpoints | edge_case | unique_key | regions | substitutions | genome_length | dataset_name | dataset_tag | cli_version |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| population_XBB | BJ.1 | novel | BJ.1,BA.2.75 | 22897-22941 | false | novel_BJ.1_BA.2.75_22897-22941 | 405-22896|BJ.1,22942-29118|BA.2.75 | A405G,T670G,C2790T,C3037T,G4184A,C4321T,C9344T,A9424G,C9534T,C9866T,C10029T,C10198T,G10447A,C10449A,C12880T,C14408T,G15451A,C15714T,C15738T,T15939C,T16342C,C17410T,T17859C,A18163G,C19955T,A20055G,C21618T,T21810C,G21987A,C22000A,C22109G,T22200A,G22577C,G22578A,G22599C,C22664A,C22674T,T22679C,C22686T,A22688G,G22775A,A22786C,G22813T,T22882G,G22895C,T22896C|BJ.1;T22942G,T23019C,T23031C,C25416T,A26275G|BA.2.75;A19326G|private | 29903 | sars-cov-2 | 2023-11-30 | 0.2.0 |
A visual representation (output/example/knockout/plots/) of the genomic composition is:
Validate
Run rebar on all populations in the dataset, and validate against the expected results.
rebar run \
--dataset-dir dataset/sars-cov-2/2023-11-30 \
--output-dir output/example/validate \
--populations "*" \
--threads 4
rebar plot \
--run-dir output/example/validate \
--annotations dataset/sars-cov-2/2023-11-30/annotations.tsv