Data Model and Permissions
This document describes a high-level overview of the proposed data model and related permissions for the IRIDA platform.
Document History
- December 12, 2013: Document creation.
- January 14, 2014: Revisions for Aaron and Josh; added to-do section.
- January 17, 2014: Recreated diagram with yed so it’s not ASCII art anymore.
- April 23, 2014: Added the concept of groups.
- April 30, 2014: Transcribed data model/permissions meeting discussion.
Authors
Background
This document describes an extension to the data model exposed by the NGS Archive, and is a proposed partial implementation of the data model described by the REST API for GMI-compliant repositories.
The data model in the NGS Archive consists of five, interrelated units:
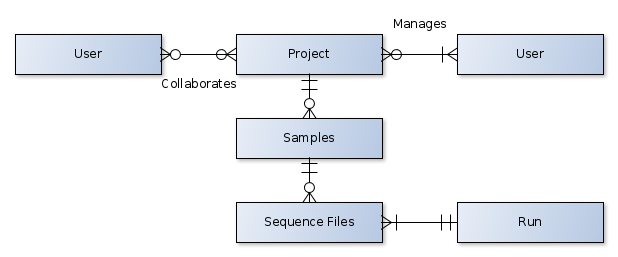
A sequence file is the atomic unit in the NGS Archive, holding a collection of metadata and read data generated by the act of sequencing an isolate.
A run is a collection of all files and metadata generated during the execution of a sequencer.
A sample (equivalently, an isolate) refers to a physical sample collected from an environment. Each sample may have one or more files representing the digital data derived from that physical sample.
A project is a collection of related samples.
The resources in the NGS Archive are protected, requiring both authentication and authorization to view any resource:
- Any user is allowed to create a project.
- The user creating the project is assigned as a manager of that project.
- Managers of a project can assign permissions to other users to read (project role of ‘user’) or write (project role of ‘manager’) to the project.
- Project permissions are transitively applied to resources contained by a project (if a user can view a project, then the user can view the samples and files contained within that project).
The proposed REST API for GMI-compliant repositories further separates a run into two resources: Run and Experiment. The experiment resource is designed to capture the laboratory methods used to prepare a sample for sequencing. The run resource captures the metadata related to the sequencing process itself (i.e., the type of sequencer used, etc.) and the files generated by the sequencer.
The data model implemented by the NGS Archive is suitable for the storage of sequencing data, but needs to be extended to support the execution of workflows.
Requirements
The IRIDA platform has several requirements not considered by the data model implemented as part of the NGS Archive:
- A method for securely storing and retrieving the results of analytical tools executed on the sequencing data stored in the platform.
- A method for specifying the public availability of data stored in the system.
- A method for storing and retrieving provenance data that can be used to accurately re-create the results of analytical tools executed as part of a workflow in the system.
Workflows
A key requirement of the IRIDA platform is ease-of-use. To that end, requirements for the execution of workflows and data selection of those workflows are:
- A user must be able to select a workflow for execution.
- A user must be presented with a collection of data to be used as input by the workflow during execution. The collection of data that the user is presented with must only be the data that they are permitted to read (i.e., the projects where the user is given read permissions, see section titled ‘User Groups and Permissions’, below).
- The user must be able to specify who (i.e., users and groups) is allowed to read the outputs produced by the execution of the workflow.
Solutions
A proposal for a data model that meets the requirements set out above includes two major components: a data project (corresponding to a study in the GMI REST API proposal and a Project in the NGS Archive) and an analysis (a container storing metadata and data related to the execution of a workflow).
Groups
A group is a logical collection of user accounts that should have the same permissions applied to a resource.
To reduce complexity, groups are only allowed to be created by administrative users (including managers) and will be visible to all users in the local system.
Data model:
- an owner (i.e., who created the group)
- a group name (unique)
- a collection of user accounts contained within the group.
Samples
Samples will exist as separate, distinct entities. In the NGS Archive, a sample was required to be contained within a parent project, however in IRIDA a sample exists outside of a project and may be referenced by zero or more projects.
A sample also corresponds to an isolate, and should have relevant epidemiological metadata attached. We derive some of the metadata definition from the NCBI BioSample project, specifically the Genome Trakr/GMI samples, that have some epidemiological metadata attached.
Data Project
- Related projects (optional)
- References to internal or external projects
- NO permissions are implied by creating a reference to another project. (An unanswered question: “If you don’t have permission to see a related project, are you allowed to see that the related project exists by having it appear in this list?”; easy answer: “Yes”, difficult answer: “No”)
- Permissions
- Project permissions are applied transitively to all sub-resources (specifically samples and sequence files)
- Individual user accounts
- Each user has a project role, one of:
- Collaborator (read-only privileges)
- Manager (user + write to metadata, create new sub-resources, add/delete read-only permissions)
- Owner (manager + add/delete write permissions)
- Each user has a project role, one of:
- Groups
- Each group has a project role (transitively applied to all users in the group), one of:
- Collaborator (read-only privileges)
- Manager (user + write to metadata, create new sub-resources, add/delete read-only permissions)
- Owner (manager + add/delete write permissions)
- Each group has a project role (transitively applied to all users in the group), one of:
- public availability (explicitly read-only access), one of:
- This project is publicly available without authentication (i.e., any client with HTTP access to the server and an appropriate URL can view the resource and all sub-resources without supplying credentials.)
- This project is publicly available with authentication (i.e., any client with HTTP access to the server, an appropriate URL and correct authentication credentials can view the resource and all sub-resources.)
- This project is private (i.e., any client with HTTP access to the server, an appropriate URL, correct authentication credentials and correct authorization can view the resource and all sub-resources.)
- Samples, a collection of isolate metadata.
- Sequence Files, a collection of sequencing reads and associated metadata.
- Analyses generated by data contained within this project.
- Questions:
- Is this all analyses that use data from this project, regardless of permissions?
- Is this all analyses that use data from this project, where only the logged-in user is permitted to read the analysis?
- Is this similar to ‘Related Projects’, where we create a link that has no implied permissions? FB: I vote for this.
- Summary question: How should this be implemented?
- Questions:
- Zero or more reference genomes related to the samples contained within this project.
Analysis
- Analyses are read-only.
- Permissions
- Individual user accounts that have read-only access to the analysis.
- Groups that have read-only access to the analysis.
- public availability, one of:
- This analysis is publicly available without authentication (i.e., any client with HTTP access to the server and an appropriate URL can view the resource and all sub-resources without supplying credentials.)
- This analysis is publicly available with authentication (i.e., any client with HTTP access to the server, an appropriate URL and correct authentication credentials can view the resource and all sub-resources.)
- This analysis is private (i.e., any client with HTTP access to the server, an appropriate URL, correct authentication credentials and correct authorization can view the resource and all sub-resources.)
- Provenance
- References to the collection of data used to generate the analysis (Implementation note by FB: “This needs to be the most granular resource used to generate the analysis; we shouldn’t store a reference to the ‘Project’ used to generate the data, rather, we should store a reference to the ‘File’ used to generate the data.”)
- Note: Read permission to the analysis explicitly does not imply read permissions to the data used to generate the analysis.
- References to the concrete version of the workflow used to generate the analysis.
- References to the collection of data used to generate the analysis (Implementation note by FB: “This needs to be the most granular resource used to generate the analysis; we shouldn’t store a reference to the ‘Project’ used to generate the data, rather, we should store a reference to the ‘File’ used to generate the data.”)
- Data (outputs)
- Granularity of outputs is left to be determined by the tools workgroup
- The reference genome used in the workflow.
An important note discussed as part of the creation of this document regarding permissions related to analyses and the data executed on by an analysis: We specified that analysis and data project have a distinct set of permissions. One problem that arises from separate permissions is that a user may have permissions to read an analysis, and may not have permissions to read the data used to generate the analysis. We identified two potential problems with this scenario:
- Reproducibility of an analysis: a user would be able to read the instructions executed on some input data (the workflow) and the output data generated by workflow execution, but would not be able to reproduce the outputs because they lack inputs.
- Reconstruction of inputs from outputs: the simplest example of this is a workflow consisting of reference mapping; the outputs of a reference mapping (a BAM file, for example) can be used to trivially reconstruct the input reads, regardless of user permissions on the data project.
Workflow Execution
The general approach to workflow execution should be:
- The user selects the type of analysis they would like to run.
- The user is provided with a list of inputs that are:
- Suitable for use as inputs by the selected analysis (i.e., requires a reference genome, sets of reads, etc.), and
- Readable by the user.
- The user is provided with the opportunity to select the permissions to be applied to an analysis (i.e., who can see the outputs?)
User Groups and Permissions
User groups are synonymous with project membership. Projects have associated permissions that relate user accounts and projects. If a user has any relationship with a project, then a user is considered to be part of the group represented by that project.
Permissions Applied to Data Projects
When assigning permissions to a data project, a project owner should be allowed to choose from:
- Individual user accounts (by specifying an e-mail address or some other unique locator for a user account),
- Groups of users on the local system.
The project owner should also be provided the option to select the access level that the user(s) should be given (i.e., one of user or manager).
If the project owner selects an individual user account, then the user account should be trivially added to the project members with the appropriate role.
If the project owner selects a group of users, then the group reference should be added to the project groups with the appropriate role. By adding a group, any changes made to the group are immediately applied to the permissions on all resources that the group is related to (for example, if a user account is removed from a group, then all permissions for all resources granted to the group are revoked for the individual user account immediately). In other words, a group is not added as a snapshot of the current group members, but rather is retained as a reference in the set of permissions applied to a resource.
The system should take the following steps when a user attempts to access a data project or data project sub-resource:
- Check to see if the data project is publicly available without authentication. If the data project is publicly available without authentication, access is permitted. If the data project is not publicly available without authentication, proceed to step 2.
- Check to see if the data project is publicly available with authentication. If the user has not provided authentication details, then permission is denied. If the user has provided authentication details, then access is permitted. If the data project is privately available, then proceed to step 3.
- The user must have provided authentication details to proceed. Check to see if the user is a member of the data project. If the user is a member of the data project, then the user has authorization to view the data project and access is permitted. If the user is not a member of the data project, then proceed to step 4.
- If the user is not a member of the data project, then iterate over the collection of group references. If the user is a member of any of those groups, access is permitted. If the user is not a member of any referred groups, access is denied.
Permissions Applied to an Analysis
When executing a workflow, a user should be provided with the opportunity to choose which other user accounts will have read privileges on the analysis outputs. The analysis creator should be able to choose from:
- Individual user accounts (by specifying an e-mail address or some other unique locator for a user account),
- Groups of users on the local system.
If the analysis creator selects an individual user account, then the user account should be given read permissions on the analysis only.
If the analysis creator selects a group of users (by selecting a project on the local system), then the selected data project should be stored as a reference.
The system should take the following steps when a user attempts to access a data project or data project sub-resource:
- Check to see if the analysis is publicly available without authentication. If the analysis is publicly available without authentication, access is permitted. If the analysis is not publicly available without authentication, proceed to step 2.
- Check to see if the analysis is publicly available with authentication. If the user has not provided authentication details, then permission is denied. If the user has provided authentication details, then access is permitted. If the analysis is privately available, then proceed to step 3.
- The user must have provided authentication details to proceed. Check to see if the user has direct read access to the analysis. If the user has direct read access to the analysis, then the user has authorization to view the analysis and access is permitted. If the user does not have direct read access to the analysis, then proceed to step 4.
- If the user does not have direct read access to the analysis, then iterate over the collection of group references. If the user is a member of any of those groups, access is permitted. If the user is not a member of any referred groups, access is denied.
Decision
This document is a proposal for implementation of two major components required by the IRIDA platform. The document will be distributed as a formal request for comments to the participants of the IRIDA platform and will change over time, including during development of the platform and related data models.
