refseq_masher: What’s in your sequence data?
refseq_masher searches your sequence data against a Mash database of 54,925 NCBI RefSeq Genomes to find what NCBI RefSeq genomes match or are contained within your sequence data.
This tutorial will run refseq_masher against a publicly available reads set.
Get tutorial reads data
Let’s get the WGS data for SRR1203042 from EBI:
Illumina MiSeq paired end sequencing; Whole genome shotgun sequencing of Salmonella enterica subsp. enterica serovar Abony str. FNW19H84 by Illumina MiSeq
Download forward and reverse reads for SRR1203042:
Initial Data
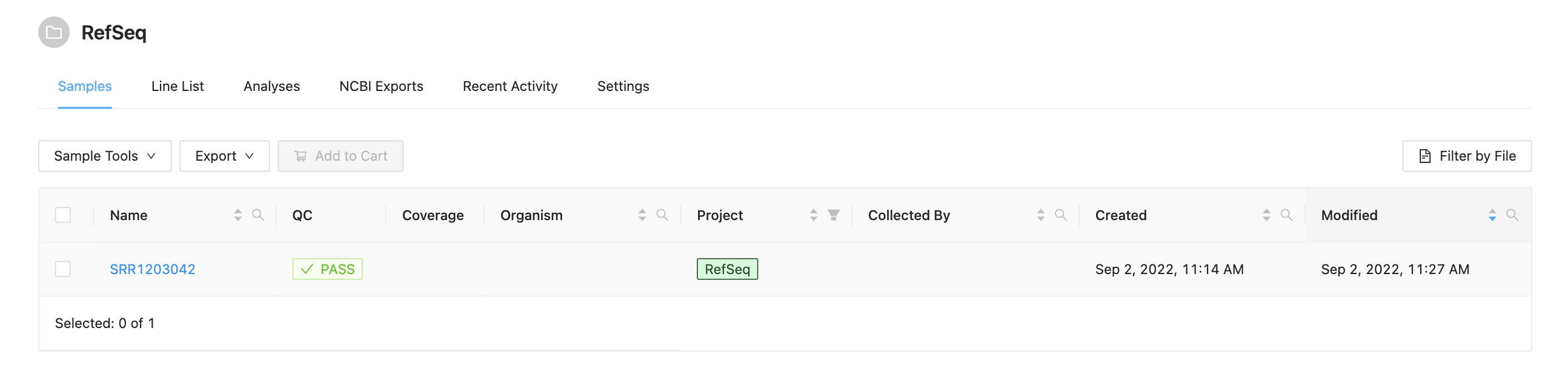
It is assumed the sequence files (forward and reverse) SRR1203042_1.fastq.gz and SRR1203042_2.fastq.gz have been uploaded into a sample as described in the Web Upload Tutorial. Before starting this tutorial you should have a project with a sample as seen below:
Add Sample to Cart
Select sample “SRR1203042” and add it to your Cart:
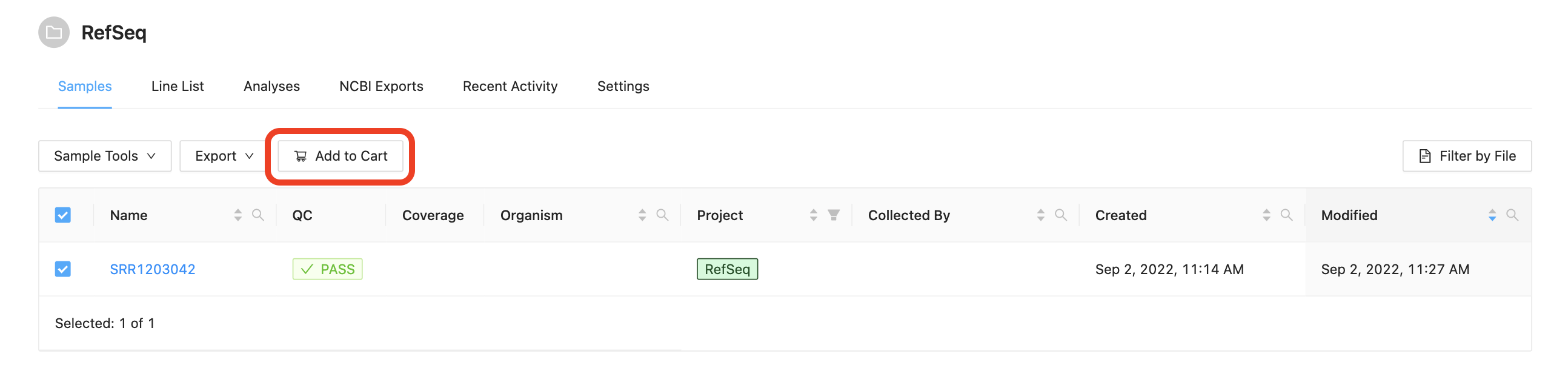
Once the samples have been added to the cart, the samples can be reviewed by clicking on the Cart button at the top.
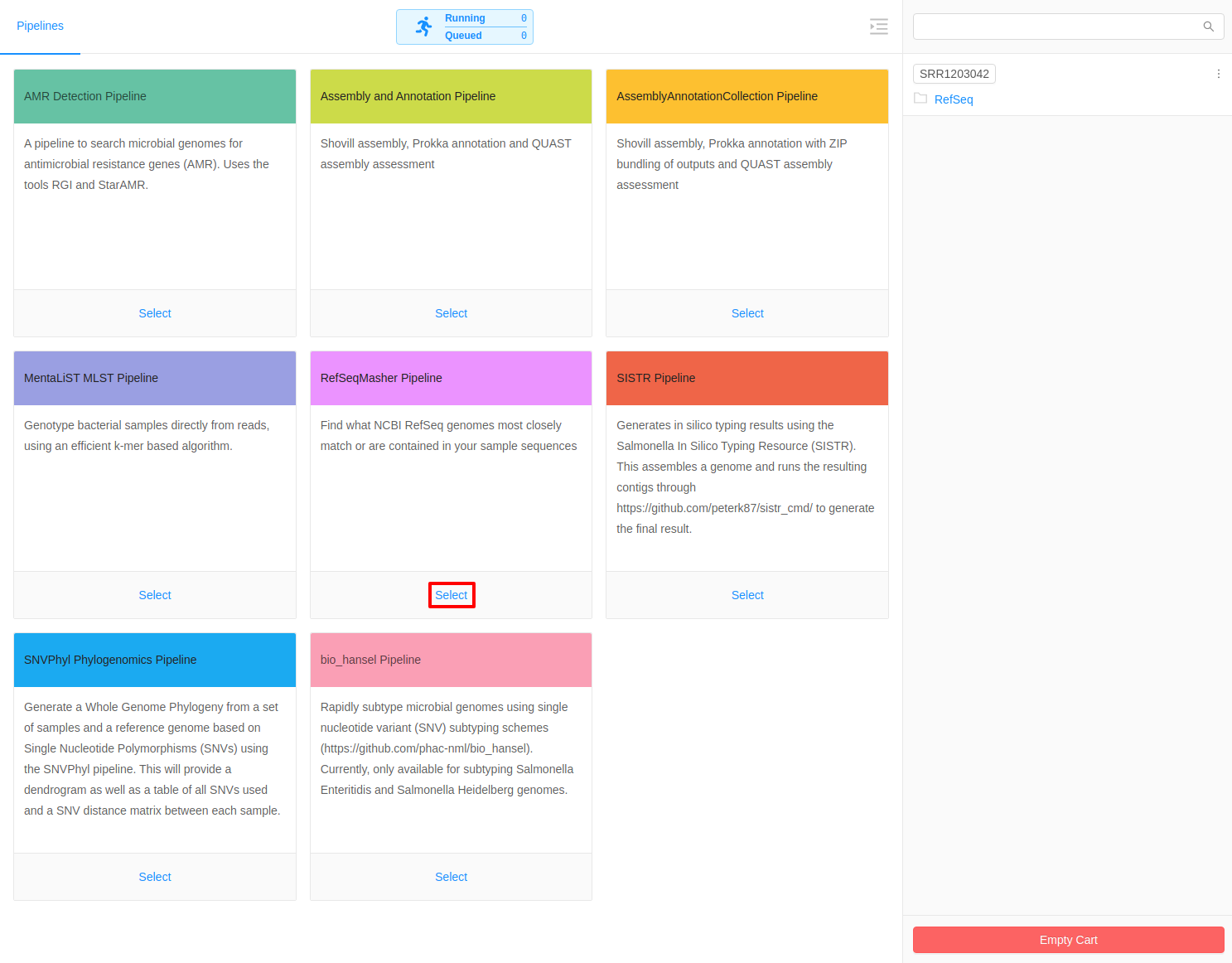
Select refseq_masher Pipeline
Once inside the cart all available pipelines will be listed in the main area of the page.
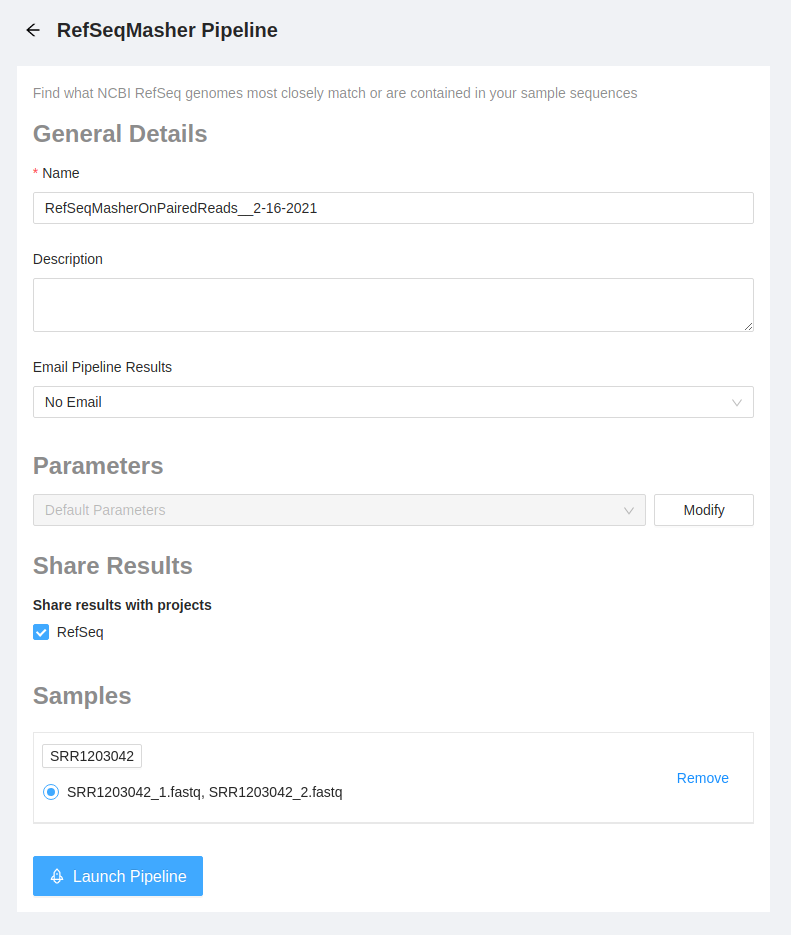
Configure and Launch refseq_masher Analysis
Modify the parameters to run refseq_masher if desired:
Click the “Launch Pipeline” button to submit the analysis.

Once the pipeline is selected, the next page provides an overview of all the input files, as well as the option to modify parameters.
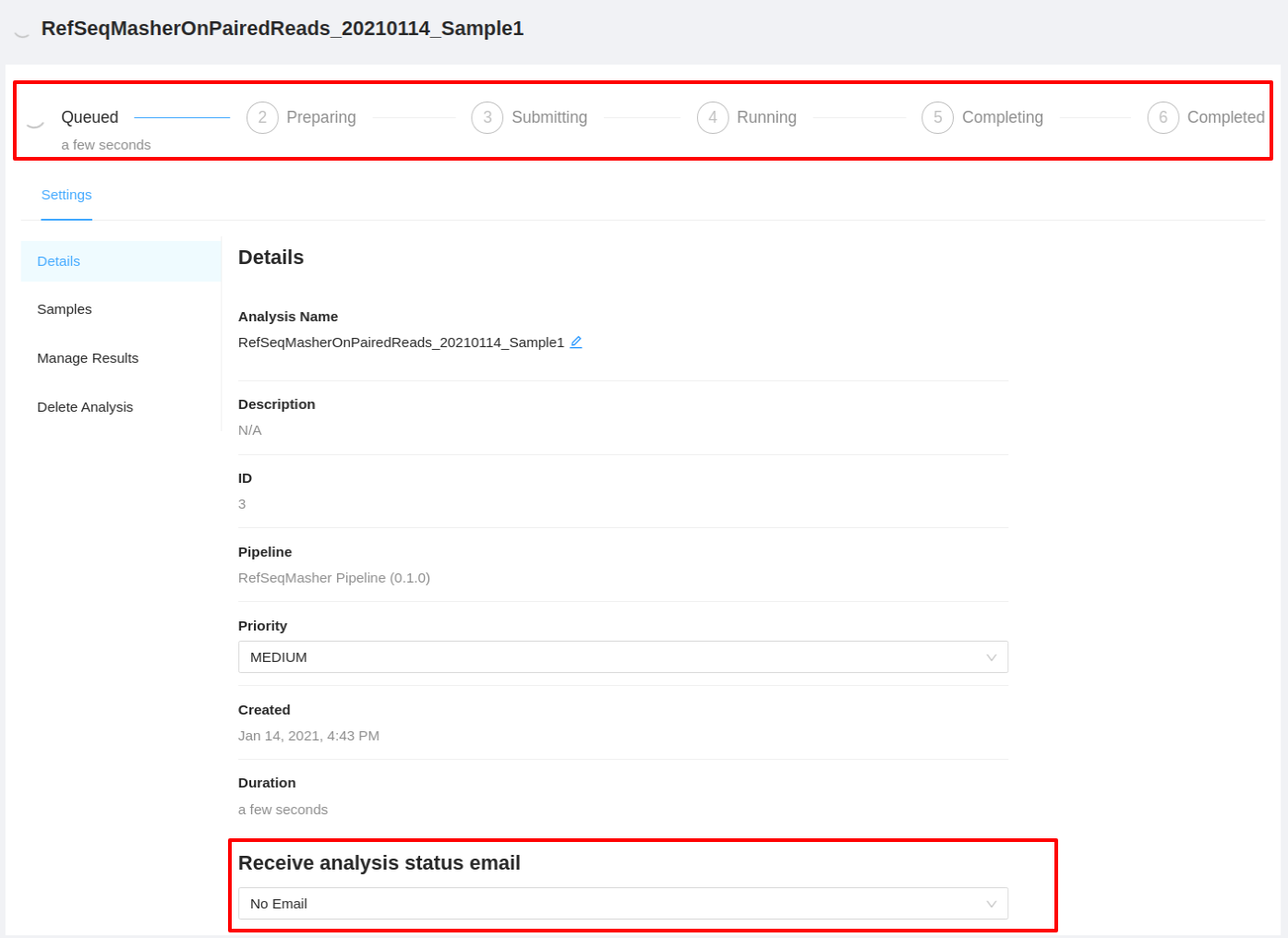
It will take a while for the RefSeqMasher analysis pipeline to complete. Along the top of the page you can check the current step of the analysis and at the bottom of the Details tab you can select if you would like to receive an email upon pipeline completion or error. The email option is only available if the analysis is not in COMPLETED or ERROR state.
When the Ref Seq Masheranalysis has completed, you will be able to view the results of the analysis. Note that not all files have an available preview and as such are not displayed in the Output File Preview but are downloaded when selecting the Download All Files button.
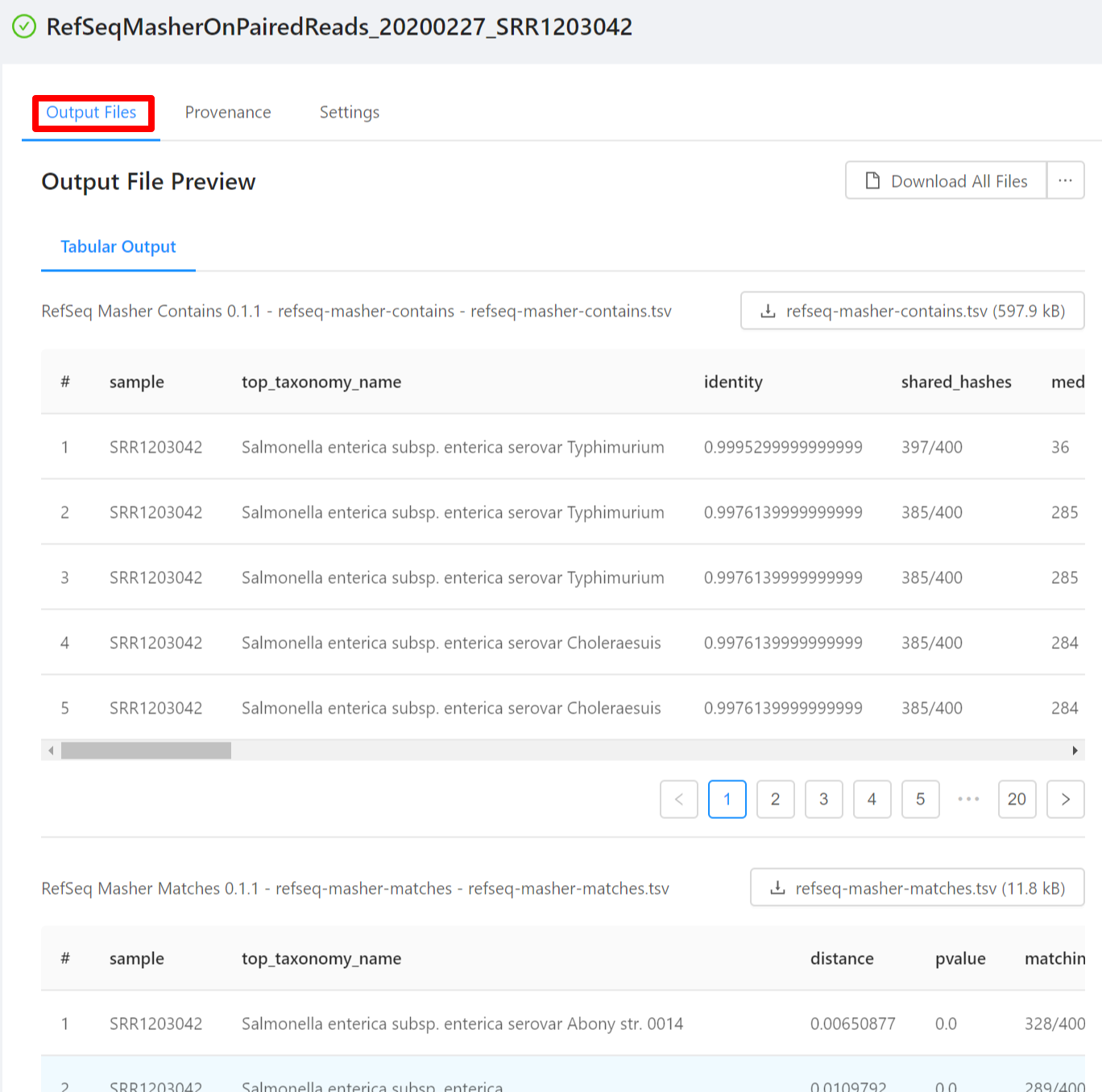
You should see 2 files under “Output File Preview”:
SRR1203042-refseq-masher-matches.tsv- Top matching NCBI Genomes to sample
SRR1203042
- Top matching NCBI Genomes to sample
SRR1203042-refseq-masher-contains.tsv- NCBI Genomes contained in sample
SRR1203042
- NCBI Genomes contained in sample
To download individual files select the … next to the Download All Files and select the file to download.
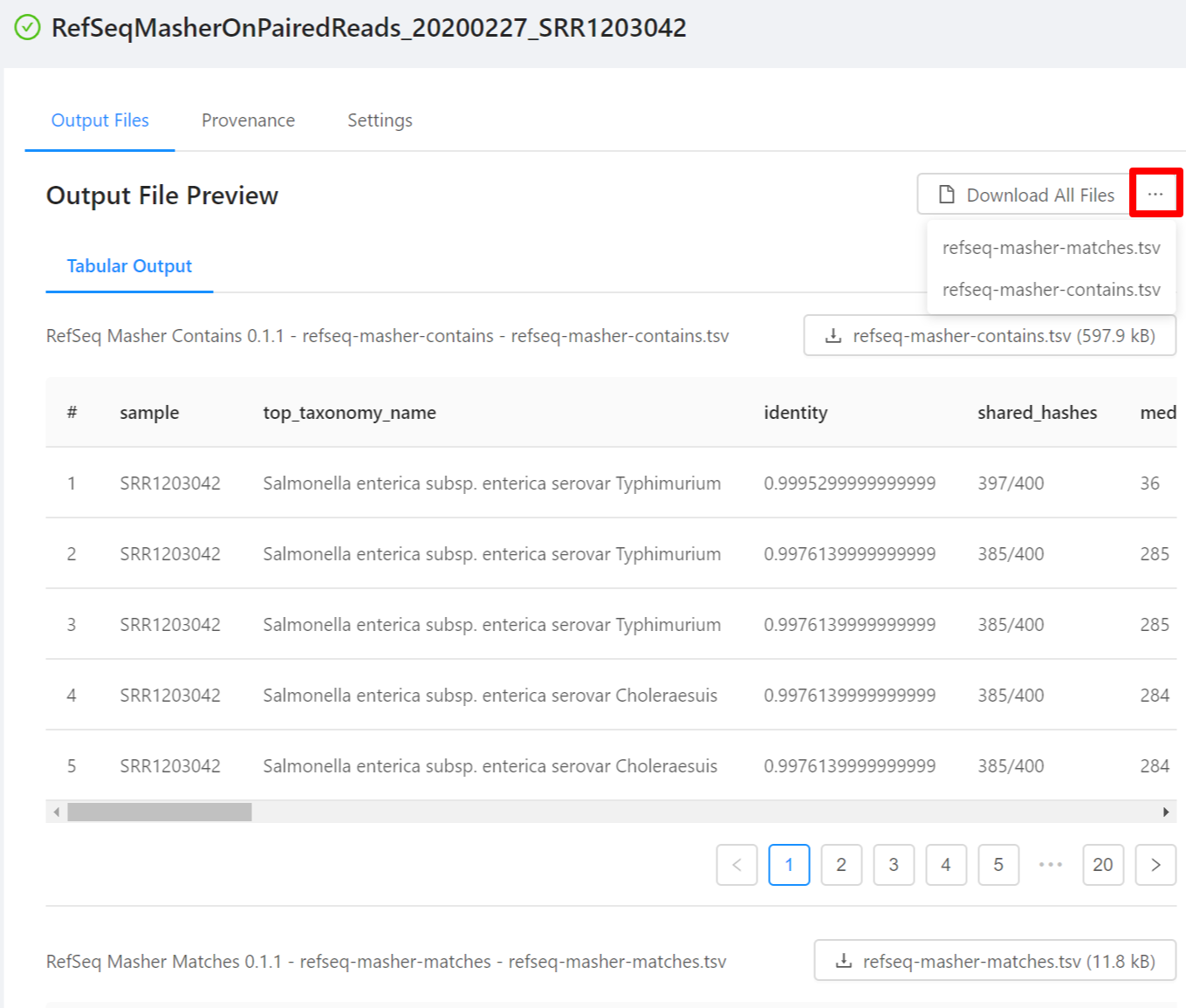
To download al the files generated by the analysis, please select the Download All Files button.
Viewing and Interpreting the Results
Matches - SRR1203042-refseq-masher-matches.tsv
Below is the top result from SRR1203042-refseq-masher-matches.tsv transposed for readability:
| Field | Value |
| sample | SRR1203042 |
| top_taxonomy_name | Salmonella enterica subsp. enterica serovar Abony str. 0014 |
| distance | 0.00650877 |
| pvalue | 0 |
| matching | 328/400 |
| full_taxonomy | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Salmonella; enterica; subsp. enterica; serovar Abony; str. 0014 |
| taxonomic_subspecies | Salmonella enterica subsp. enterica |
| taxonomic_species | Salmonella enterica |
| taxonomic_genus | Salmonella |
| taxonomic_family | Enterobacteriaceae |
| taxonomic_order | Enterobacterales |
| taxonomic_class | Gammaproteobacteria |
| taxonomic_phylum | Proteobacteria |
| taxonomic_superkingdom | Bacteria |
| subspecies | enterica |
| serovar | Abony |
| plasmid | |
| bioproject | PRJNA224116 |
| biosample | SAMN01823751 |
| taxid | 1029983 |
| assembly_accession | GCF_000487615.2 |
| match_id | ./rcn/refseq-NZ-1029983-PRJNA224116-SAMN01823751-GCF_000487615.2-.-Salmonella_enterica_subsp._enterica_serovar_Abony_str._0014.fna |
Given that our sample, SRR1203042, was from an Illumina MiSeq sequencing run of strain Salmonella enterica subsp. enterica serovar Abony str. FNW19H84, the top refseq_masher result confirms that the WGS is Salmonella enterica subsp. enterica serovar Abony.
For more info on interpreting refseq_masher matches results, see refseq_masher matches documentation
Contains - SRR1203042-refseq-masher-contains.tsv
If a read set potentially has multiple genomes, it can be “screened” against the database to estimate how well each genome is contained in the read set.
- Mash Screen tutorial
Below are the first 5 rows and columns of SRR1203042-refseq-masher-contains.tsv:
| sample | top_taxonomy_name | identity | shared_hashes | median_multiplicity |
| SRR1203042 | Salmonella enterica subsp. enterica serovar Typhimurium | 0.99953 | 397/400 | 36 |
| SRR1203042 | Salmonella enterica subsp. enterica serovar Typhimurium | 0.997614 | 385/400 | 285 |
| SRR1203042 | Salmonella enterica subsp. enterica serovar Typhimurium | 0.997614 | 385/400 | 285 |
| SRR1203042 | Salmonella enterica subsp. enterica serovar Choleraesuis | 0.997614 | 385/400 | 284 |
| SRR1203042 | Salmonella enterica subsp. enterica serovar Choleraesuis | 0.997614 | 385/400 | 284 |
We can see that SRR1203042 mostly contains k-mers belonging to Salmonella enterica subsp. enterica NCBI RefSeq Genomes.
About some of the fields in SRR1203042-refseq-masher-contains.tsv:
sample- Your sample nameidentity- Proportion of identical hashes or k-mers between your sample and an NCBI RefSeq Genome in the Mash Sketch databaseshared_hashes- Number of hashes shared between your sample and an NCBI RefSeq Genome in the Mash Sketch databasemedian_multiplicity- “median multiplicity is computed for shared hashes, based on the number of observations of those hashes within the pool” (frommash screen -hwith Mash v2.0)
For more info on interpreting refseq_masher contains results, see refseq_masher contains documentation and the Mash Screen documentation.
Viewing Provenance Information
To view the pipeline provenance information, please select the Provenance tab.
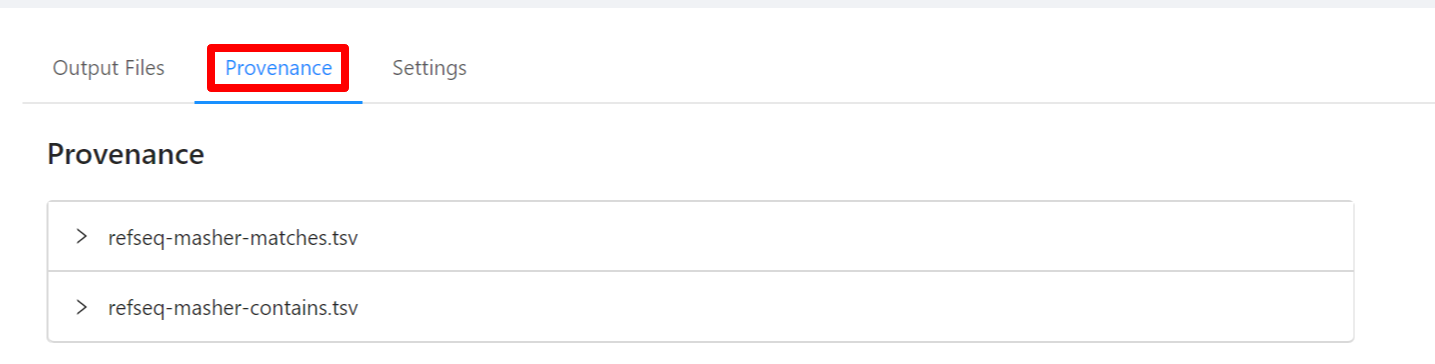
The provenance is displayed on a per file basis. Clicking on refseq-masher-matches.tsv file will display it’s provenance. Expanding each tool will display the parameters that the tool was executed with.
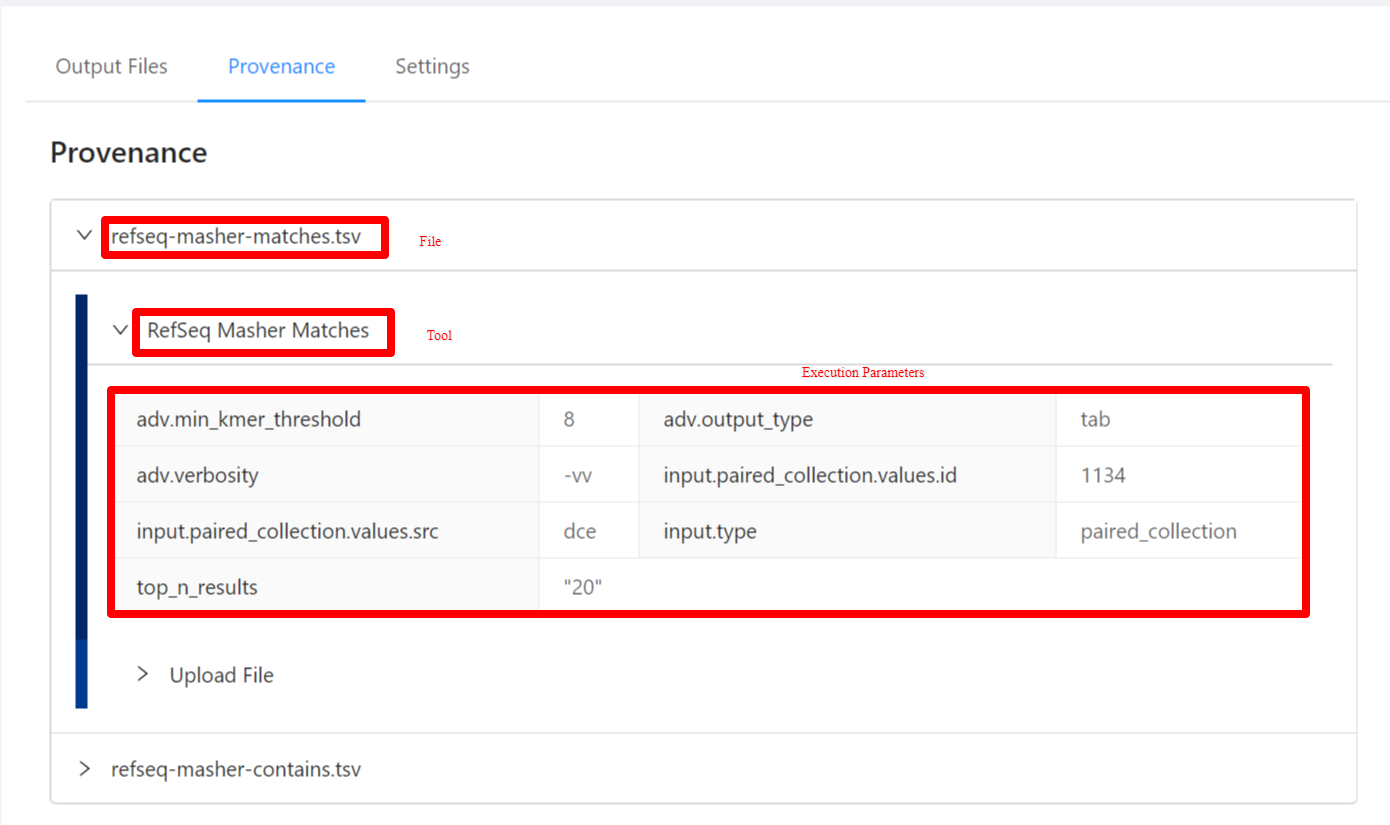
Viewing Pipeline Details
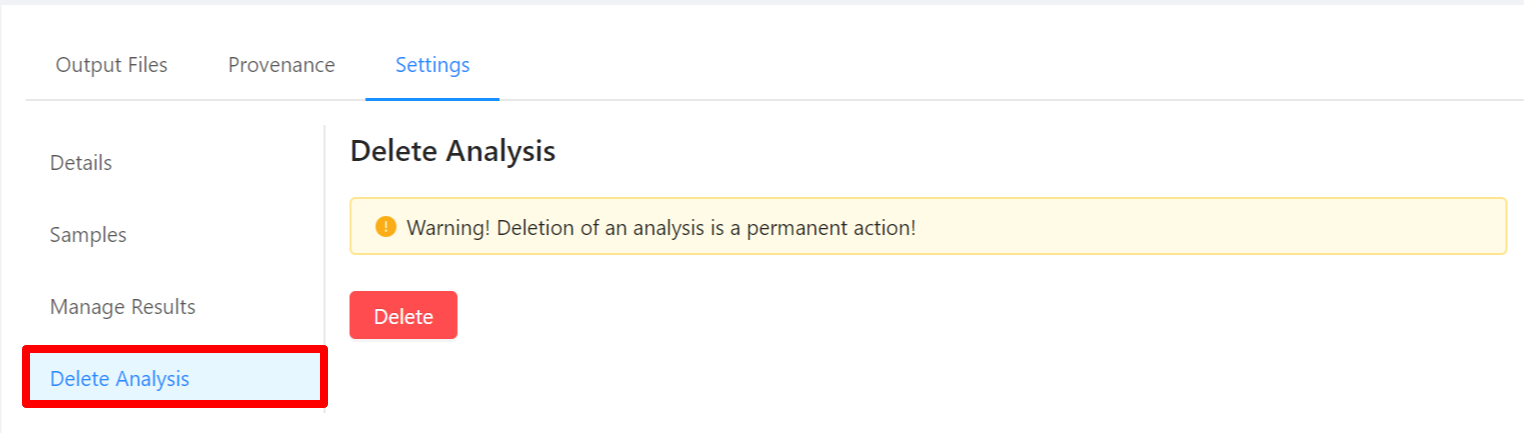
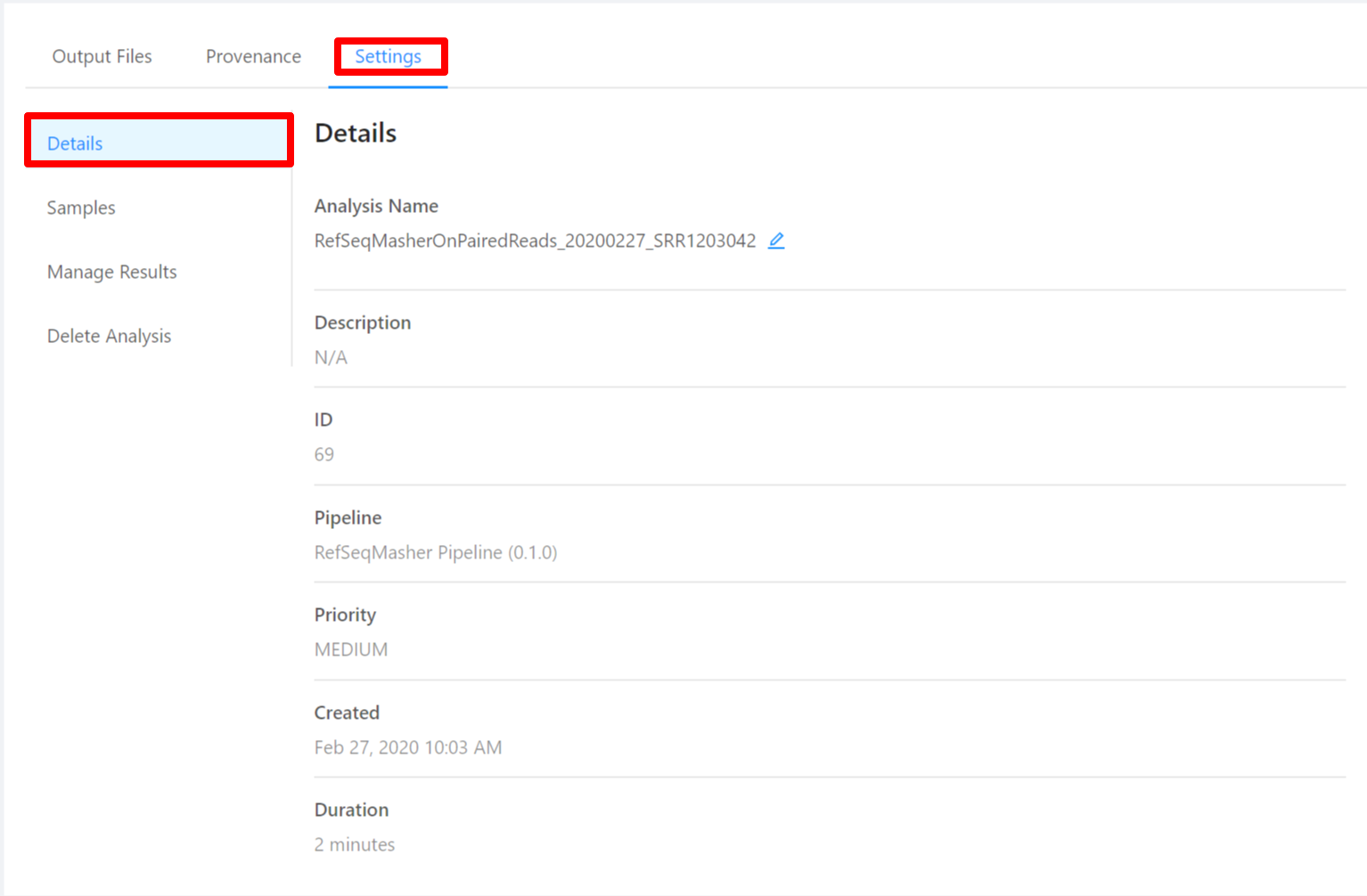
To view analysis details, please select the Settings tab. From here you can view the analysis name, analysis description, analysis id, pipeline and pipeline version used by the analysis, analysis priority, when the analysis was created, and duration of the analysis.
To edit an analysis name, please select the Pencil icon next to the analysis name. Once you have edited the analysis name, pressing the ENTER key on your keyboard or clicking anywhere outside of the text box will update the name. To cancel editing a name you can either hit the ESC key on your keyboard or if the name has not been changed you can also click anywhere outside of the text box.
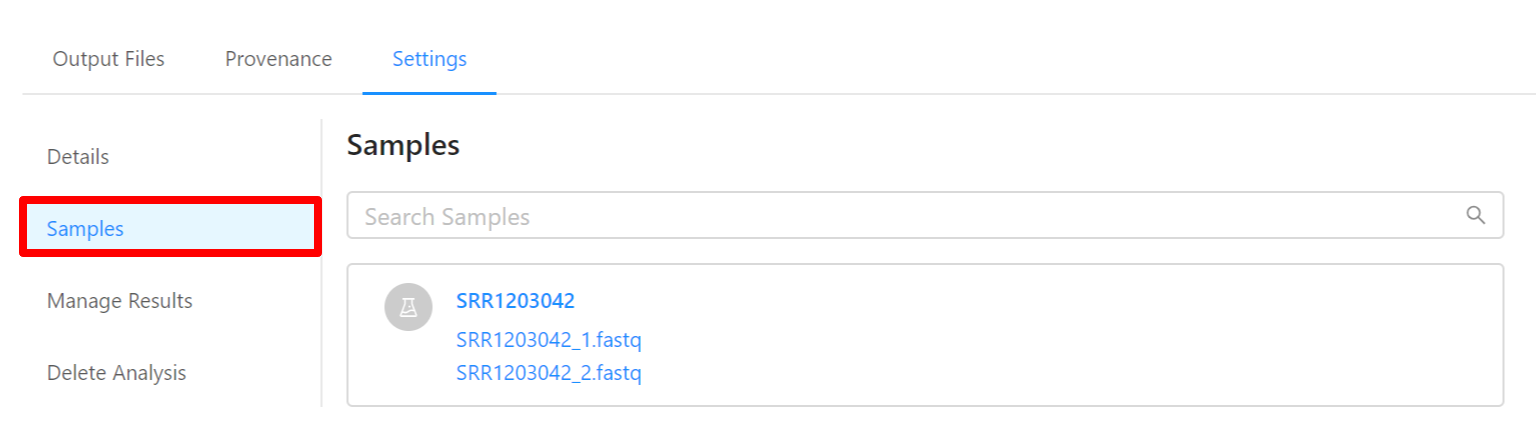
To view samples used by the analysis, please select the Samples tab.
To share analysis results with other projects, please select the Manage Results tab.
To delete an analysis, please select the Delete Analysis tab.