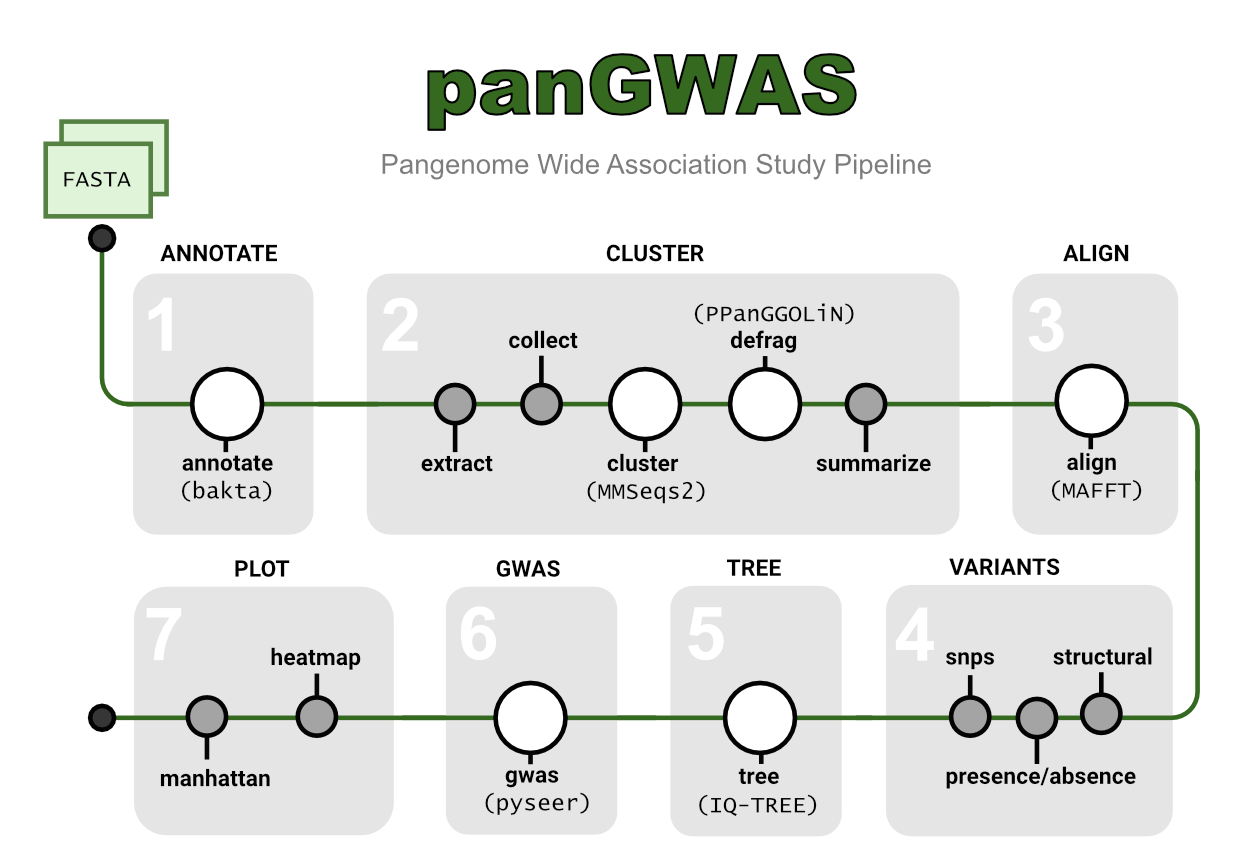
Manual
Annotate
- annotate: Annotate genomic assemblies with bakta.
Cluster
- extract: Extract sequences and annotations from GFF files.
- collect: Collect extracted sequences from multiple samples into one file.
- cluster: Cluster nucleotide sequences with mmseqs.
- defrag: Defrag clusters by associating fragments with their parent cluster.
- summarize: Summarize clusters according to their annotations.
Align
- align: Align clusters using mafft and create a pangenome alignment.
Variants
- structural: Extract structural variants from cluster alignments.
- snps: Extract SNPs from a pangenome alignment.
- presence_absence: Extract presence absence of clusters.
Tree
- tree: Estimate a maximum-likelihood tree with IQ-TREE.
GWAS
- gwas: Run genome-wide association study (GWAS) tests with pyseer.
Plot
- manhattan: Plot the distribution of variant p-values across the genome.
- heatmap: Plot a heatmap of variants alongside a tree.
Utility
- root_tree: Root tree on outgroup taxa.
- binarize: Convert a categorical column to multiple binary (0/1) columns.
- table_to_rtab: Convert a TSV/CSV table to an Rtab file based on regex filters.
- vcf_to_rtab: Convert a VCF file to an Rtab file.
Overview