Tutorial 03 - Pyseer Tutorial
This tutorial automates and reproduces the results from the penicillin resistance GWAS written by the pyseer authors.
Download the tutorial data.
git clone --depth 1 https://github.com/phac-nml/pangwas.git cd pangwasDecompress the data.
gunzip data/tutorial_core/snps.Rtab.gz gunzip data/tutorial_pangenome/variants.Rtab.gz gunzip data/tutorial_pangenome/clusters.tsv.gzRun the core genome GWAS on pencillin resistance.
nextflow run phac-nml/pangwas -profile tutorial_coreOpen up the manhattan plot in Edge or Firefox.
- Path:
results/tutorial_core/manhattan/penicillin/penicillin.plot.svg - Hover your mouse over variants to see contextual information.
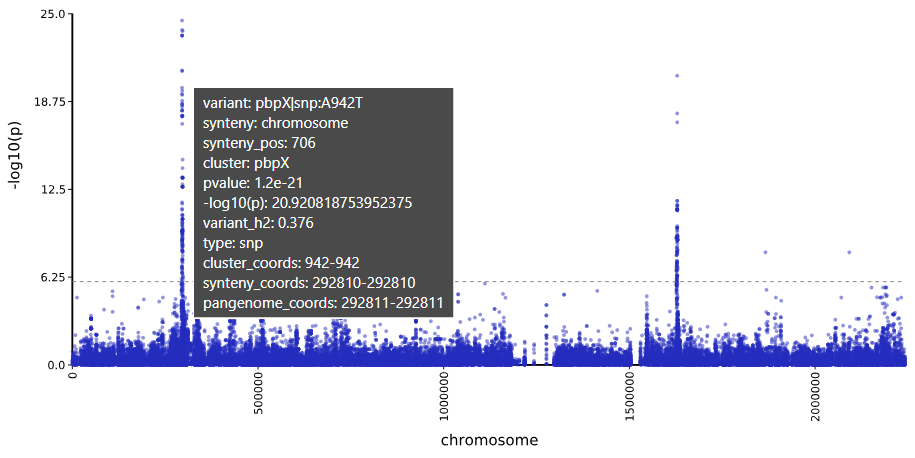
- Path:
Run the pangenome GWAS on penicillin resistance.
nextflow run phac-nml/pangwas -profile tutorial_pangenome
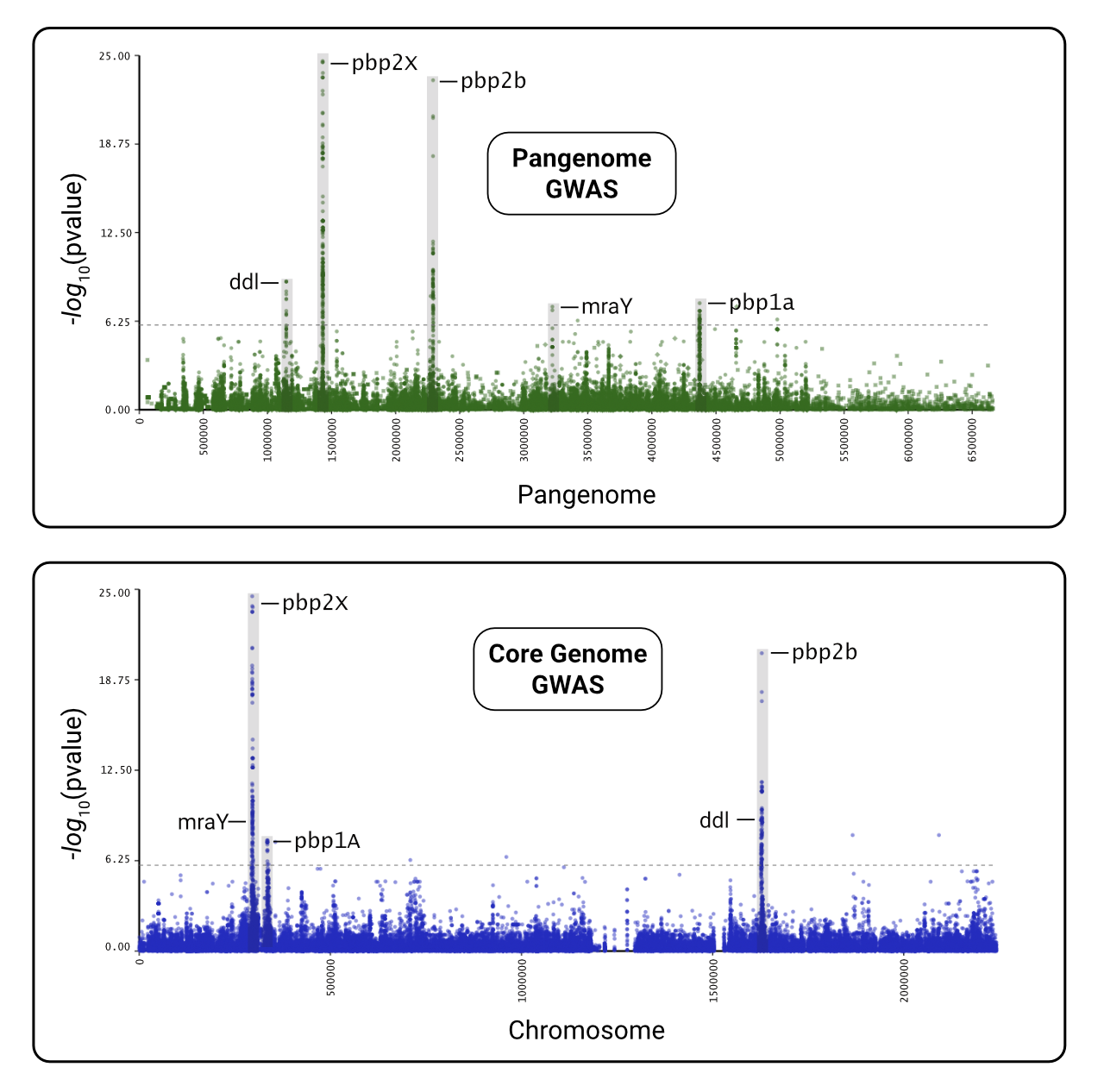
Penicillin resistance is primarily controlled by core genome genes, and we can see that the major genes are identical between a pangenome and core genome GWAS.
The following image was made by importing the raw SVG plots into a vector graphics program (ex. Affinity) and adding text and box overlays to highlight the significant variants.