Exporting data to NCBI’s Sequence Read Archive
IRIDA can assist in uploading sequence files to NCBI’s Sequence Read Archive. IRIDA requires that BioProjects and BioSamples be created before uploading, and will assign uploaded sequence files to the given BioProject and BioSample identifiers. More information about the metadata which must be entered during the upload process can be found at NCBI Submission Quick Start Guide.
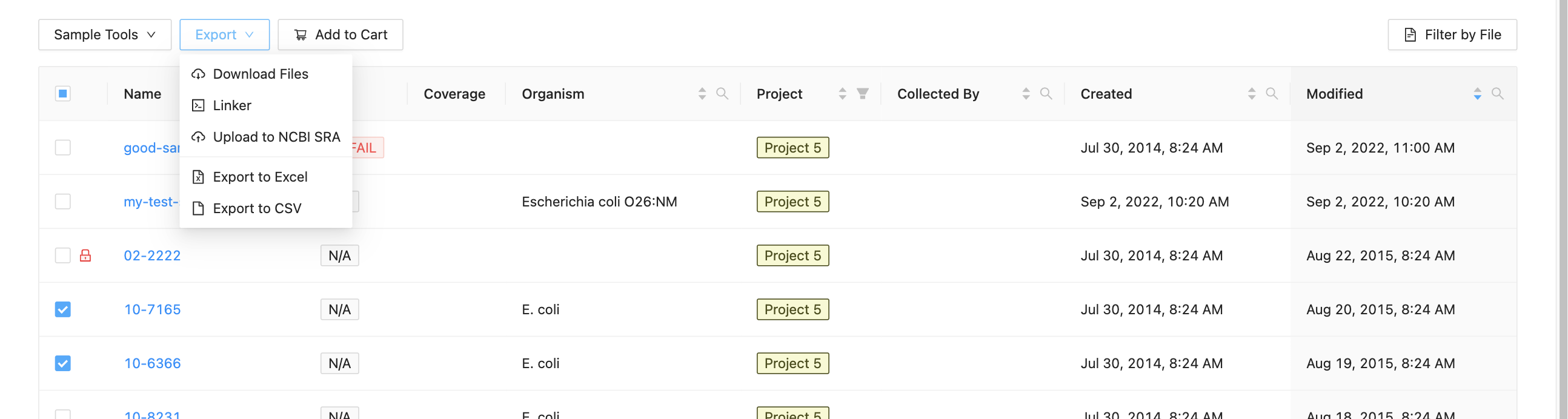
To begin submitting sequence files, select which samples you want to upload from the project samples page, then click the Export and Upload to NCBI SRA button.
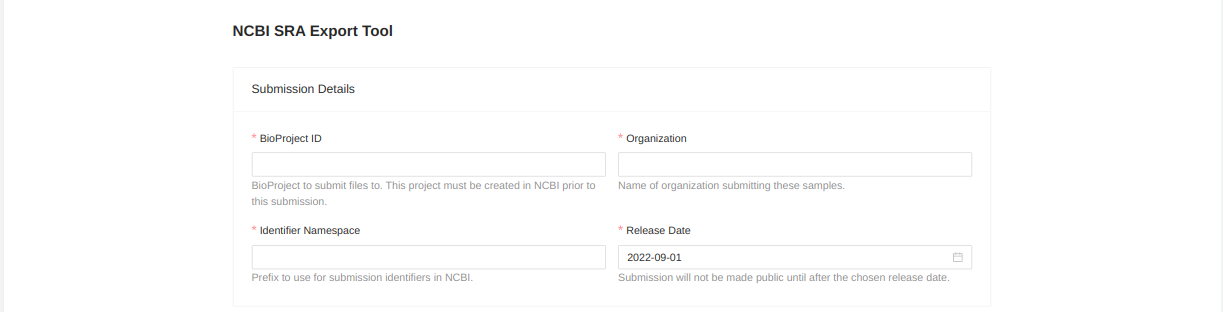
You will be forwarded to a page where you must enter metadata about the uploaded files. Start by entering information about the upload:
- BioProject ID - BioProject to submit files to. This project must be created in NCBI prior to this submission.
- Organization - Name of organization submitting these samples.
- Identifier Namespace - Prefix to use for submission identifiers in NCBI. This prefix will be used to assign upload identifiers in the SRA but may not be visible in the uploaded files.
- Release Date - Submission won’t be public until after the chosen release date.
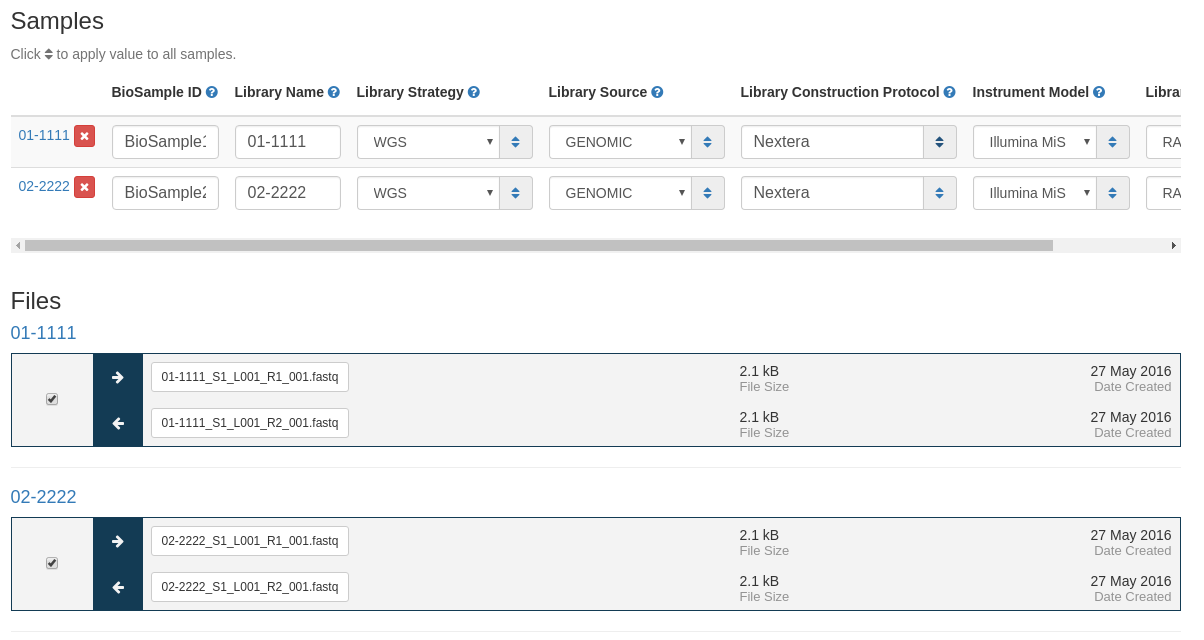
Next you must fill in information about the samples to be uploaded.
- BioSample ID - NCBI BioSample to add files to. This sample must be created in NCBI prior to this submission.
- Library Name - The submitters name for this library.
- Library Strategy - Sequencing technique intended for this library.
- Library Source - The type of source material that is being sequenced.
- Library Construction Protocol - Free form text describing the protocol by which the sequencing library was constructed.
- Instrument Model - The sequencing platform used to produce the data.
- Library Selection - Whether any method was used to select for or against, enrich, or screen the material being sequenced.
Default values for all samples can be set by clicking on the “Default Sample Settings” and setting the values for Library Strategy, Library Source, Library Construction Protocol, Instrument Model, Library Selection. You can set any number of these fields that you want, and the values can be overwritten directly within each sample.

Once all fields and files are selected for a sample, the label will change from “MISSING DATA” to “VALID” indicating that the sample is ready for submission. All samples must be valid before submitting.

After entering this metadata you can select which files should be uploaded from each sample. Only files selected with checkboxes will be uploaded to NCBI.
Click the Submit at the bottom of the page when the information is complete.
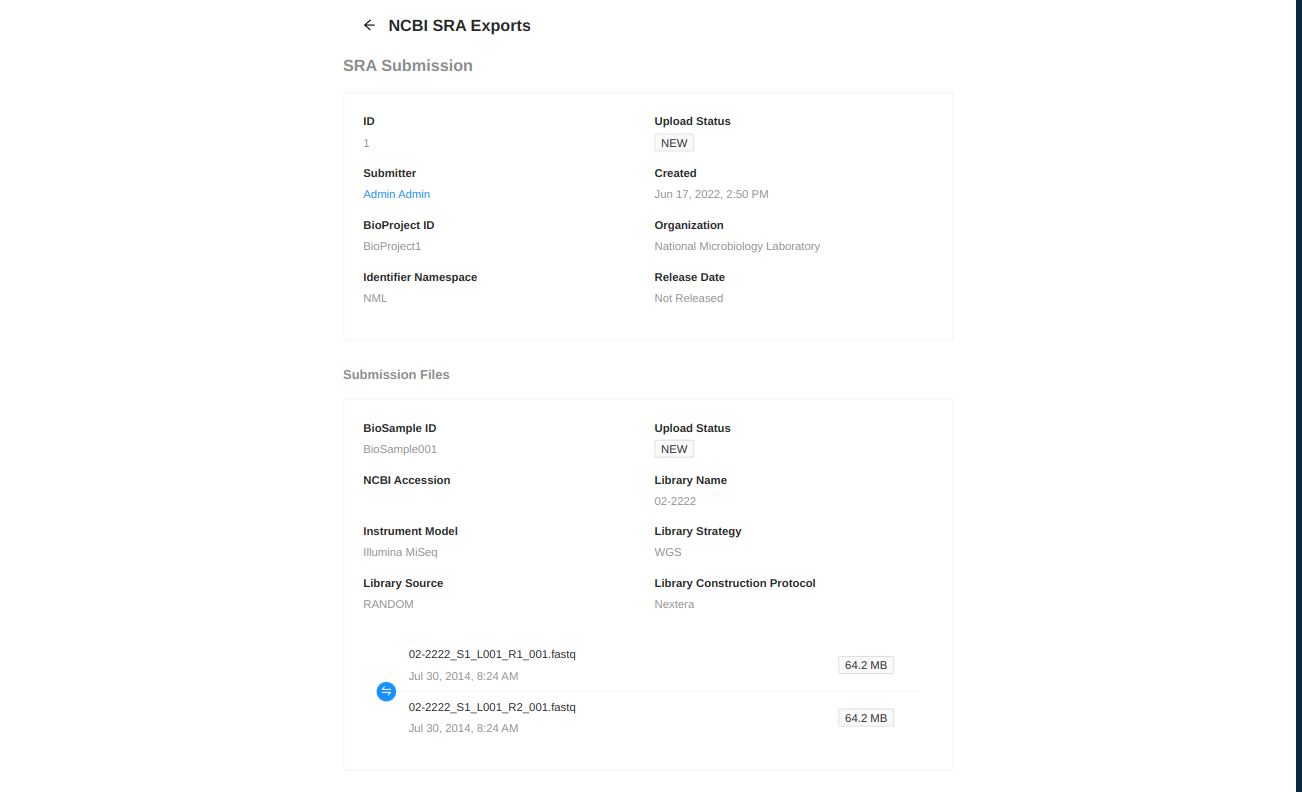
After submitting you will be redirected to a page showing the information you have entered for the upload and the status of the upload. IRIDA will periodically check the status of uploads in the SRA and update their status as necessary. After NCBI has assigned an accession number to your upload it will be displayed on this page.