rebar
rebar is a REcombination BARcode detector!
Why rebar?
-
rebardetects and visualizes genomic recombination.It follows the PHA4GE Guidance for Detecting and Characterizing SARS-CoV-2 Recombinants which outlines three steps:
- Assess the genomic evidence for recombination.
- Identify the breakpoint coordinates and parental regions.
- Classify sequences as designated or novel recombinant lineages.
-
rebarpeforms generalized clade assignment.While specifically designed for recombinants,
rebarworks on non-recombinants tool! It will report a sequence’s closest known match in the dataset, as well any mutation conflicts that were observed. The linelist and visual outputs can be used to detect novel variants, such as the SARS-CoV-2 pango-designation process. -
rebaris for exploring hypotheses.The recombination search can be customized to test your hypotheses about which parents and genomic regions are recombining. If that sounds overwhelming, you can always just use the pre-configured datasets (ex. SARS-CoV-2) that are validated against known recombinants.
Install
rebar is a standalone binary file, we recommend conda or direct download.
conda install -c bioconda rebar
- Please see the install docs for Windows, macOS, Docker, Singularity, and Conda.
- Please see the compile docs for those interested in source compilation.
Usage
Custom Dataset
A small, test dataset (toy1) serves as a template for creating custom datasets, and for easer visualization of the method and output.
rebar dataset download --name toy1 --tag custom --output-dir dataset/toy1
rebar run --dataset-dir dataset/toy1 --populations "*" --mask 0,0 --min-length 3 --output-dir output/toy1
rebar plot --run-dir output/toy1 --annotations dataset/toy1/annotations.tsv
SARS-CoV-2
Download a SARS-CoV-2 dataset, version-controlled to the date 2023-11-30 (try any date!).
rebar dataset download --name sars-cov-2 --tag 2023-11-30 --output-dir dataset/sars-cov-2/2023-11-30
rebar run --dataset-dir dataset/sars-cov-2/2023-11-30 --populations "AY.4.2*,BA.5.2,XBC.1.6*,XBB.1.5.1,XBL" --output-dir output/sars-cov-2
rebar plot --run-dir output/sars-cov-2 --annotations dataset/sars-cov-2/2023-11-30/annotations.tsv
Other
Please see the examples docs for more tutorials including:
- Using your own alignment of genomes as input.
- Testing specific parent combinations.
- Performing a ‘knockout’ experiment.
- Validating all populations in a dataset.
Please see the dataset and run docs for more methodology.
Output
Linelist
A linelist summary of results (ex. output/toy1/linelist.tsv).
| strain | validate | validate_details | population | recombinant | parents | breakpoints | edge_case | unique_key | regions | genome_length | dataset_name | dataset_tag | cli_version |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| population_A | pass | A | false | 20 | toy1 | custom | 0.2.0 | ||||||
| population_B | pass | B | false | 20 | toy1 | custom | 0.2.0 | ||||||
| population_C | pass | C | false | 20 | toy1 | custom | 0.2.0 | ||||||
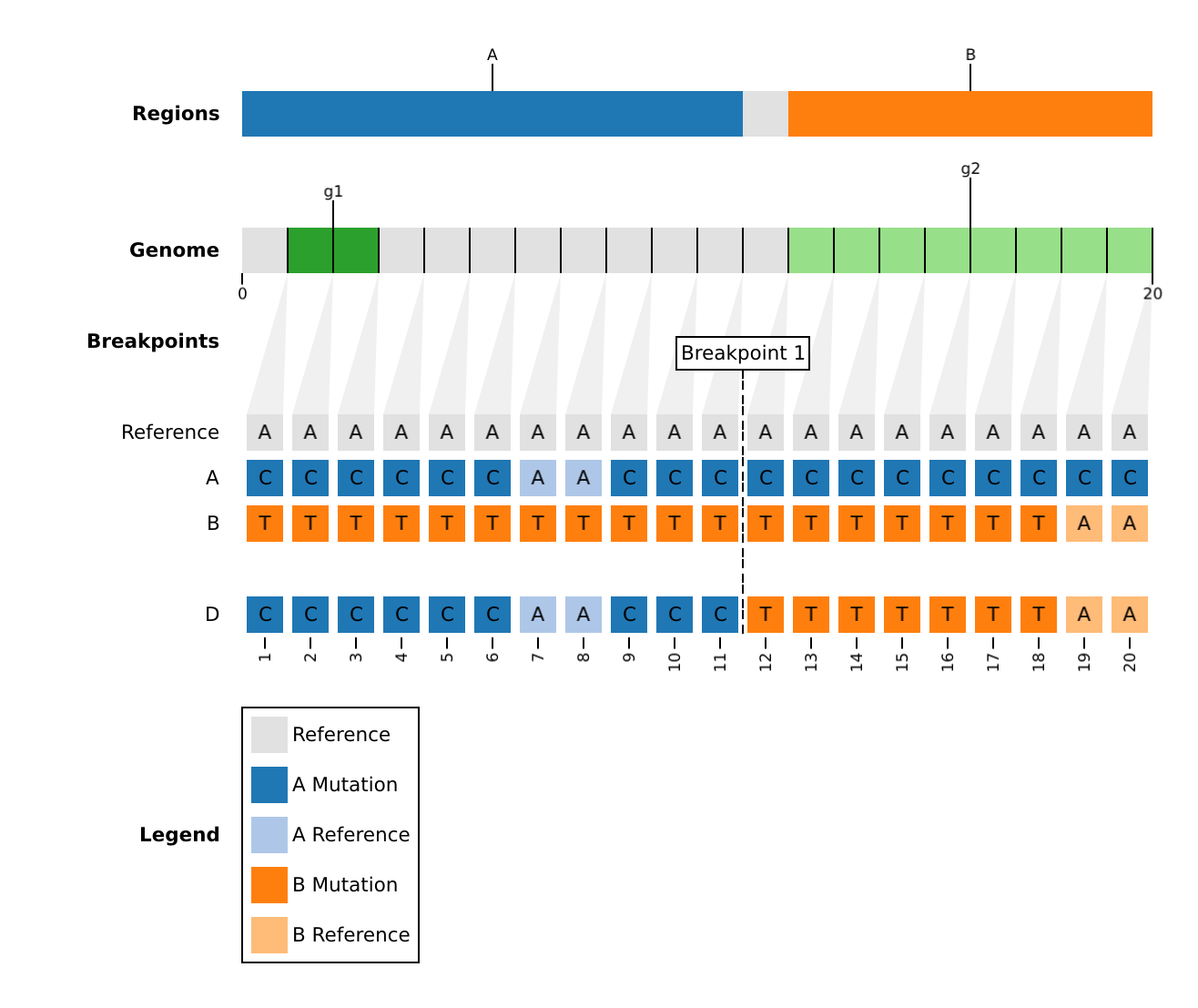
| population_D | pass | D | D | A,B | 12-12 | false | D_A_B_12-12 | 1-11|A,12-20|B | 20 | toy1 | custom | 0.2.0 | |
| population_E | pass | E | E | C,D | 4-4 | false | E_C_D_4-4 | 1-3|C,4-20|D | 20 | toy1 | custom | 0.2.0 |
Plots
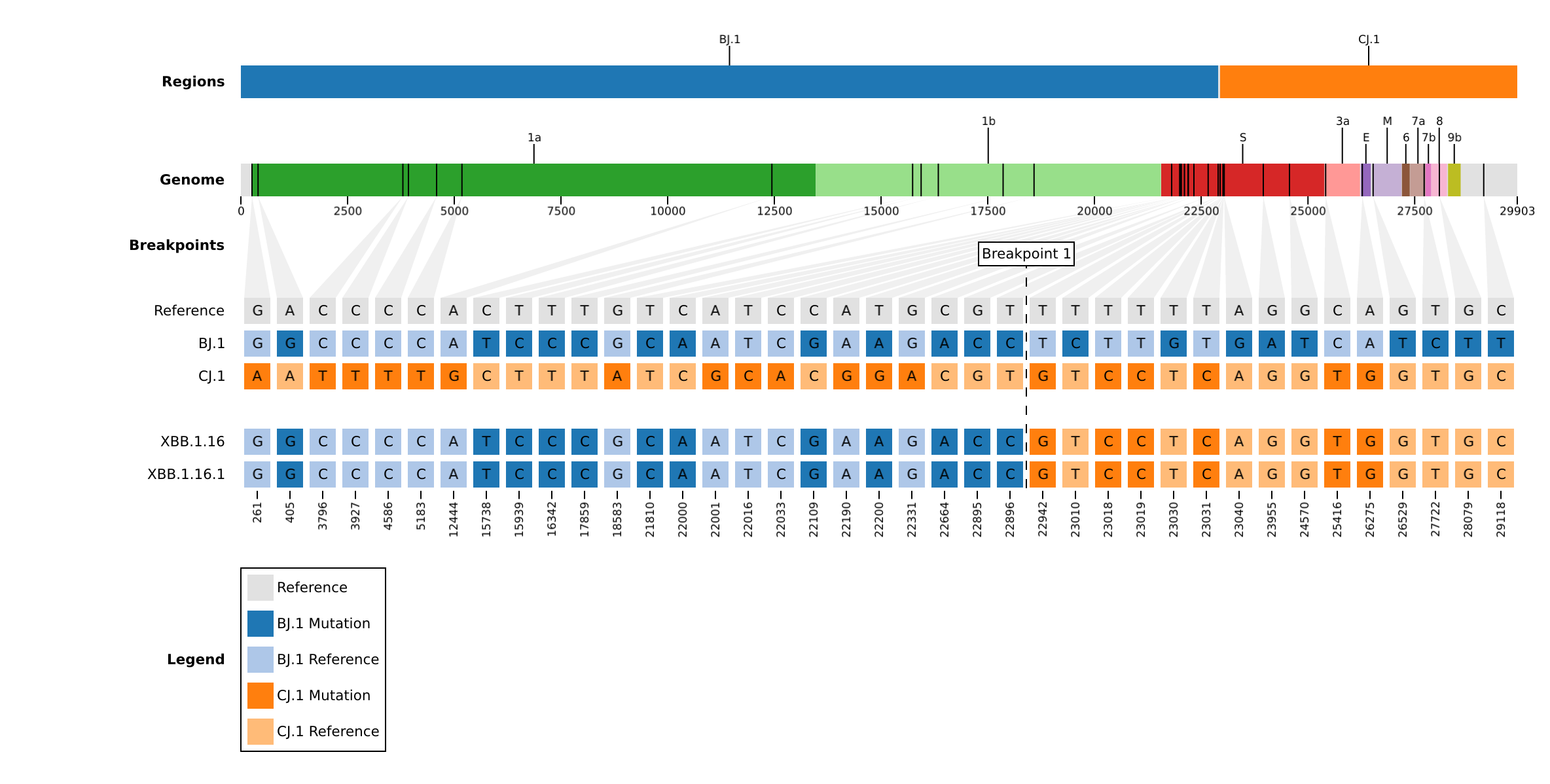
A visualization of substitutions, parental origins, and breakpoints (ex. output/toy1/plots/).
Barcodes
The discriminating sites with mutations between samples and their parents (ex. output/toy1/barcodes/).
| coord | origin | Reference | A | B | population_D |
|---|---|---|---|---|---|
| 1 | A | A | C | T | C |
| 2 | A | A | C | T | C |
| 3 | A | A | C | T | C |
| 4 | A | A | C | T | C |
| 5 | A | A | C | T | C |
| … | … | … | … | … | … |
Credits
rebar is built and maintained by Katherine Eaton at the National Microbiology Laboratory (NML) of the Public Health Agency of Canada (PHAC).
This project follows the all-contributors specification (emoji key). Contributions of any kind welcome!
Katherine Eaton 💻 📖 🎨 🤔 🚇 🚧 |
Special thanks go to the following people, who are instrumental to the design and data sources in rebar:
- Lena Schimmel (@lenaschimmel) for the original concept of a barcode-scanning recombinant detector with sc2rf.
- Cornelius Roemer (@corneliusroemer) for the designated lineages, consensus sequences, and Nextclade barcodes.
- Josh Levy (@joshuailevy) and the Andersen Lab for the UShER barcodes from Freyja.
- Richard Neher (@rneher) and the Neher Lab for python package structure, specifically treetime.
|
Lena Schimmel 🤔 |
Cornelius Roemer 🔣 🔣 🔣 |
Josh Levy 🔣 |
Richard Neher 🤔 |
Thanks go to the following people, who participated in the development of rebar and ncov-recombinant:




