Exporting data to the command-line
The IRIDA package comes with a Linux command-line utility for linking to files in your current working directory. If you are working on a Linux workstation, we strongly encourage you to use the command-line utility for working with the sequencing data stored in IRIDA.
Start by selecting the samples that you want to export to the command-line, clicking on the “Export” button just above the samples list and clicking “Command-line Linker”:
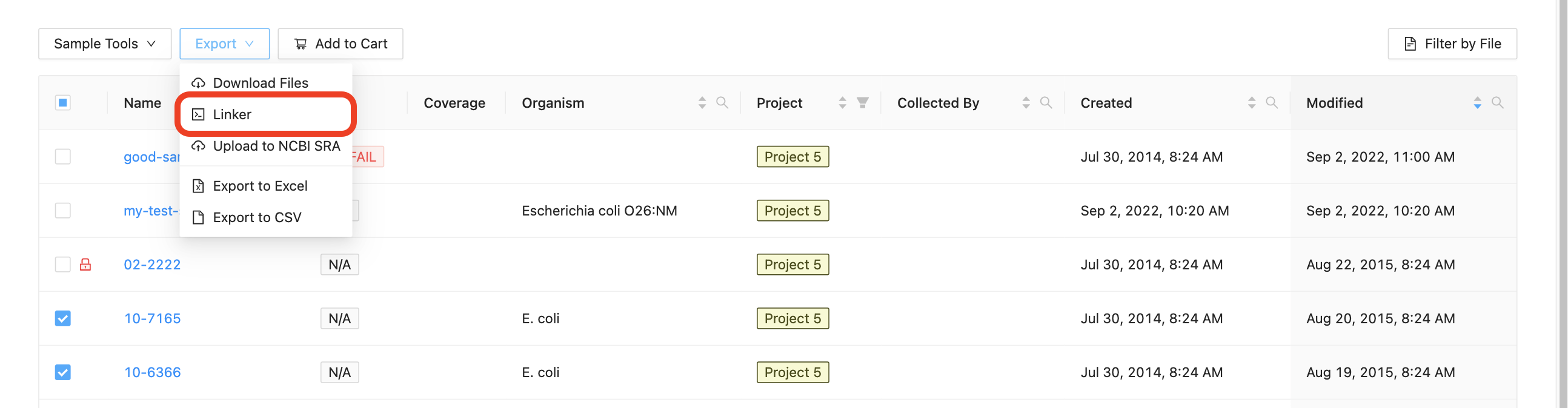
The dialog that appears will provide you with a command that you can copy and paste into a terminal window. You may select “Sequence Files” to get raw sequencing data, or “Assemblies” to get assembled data, or both.
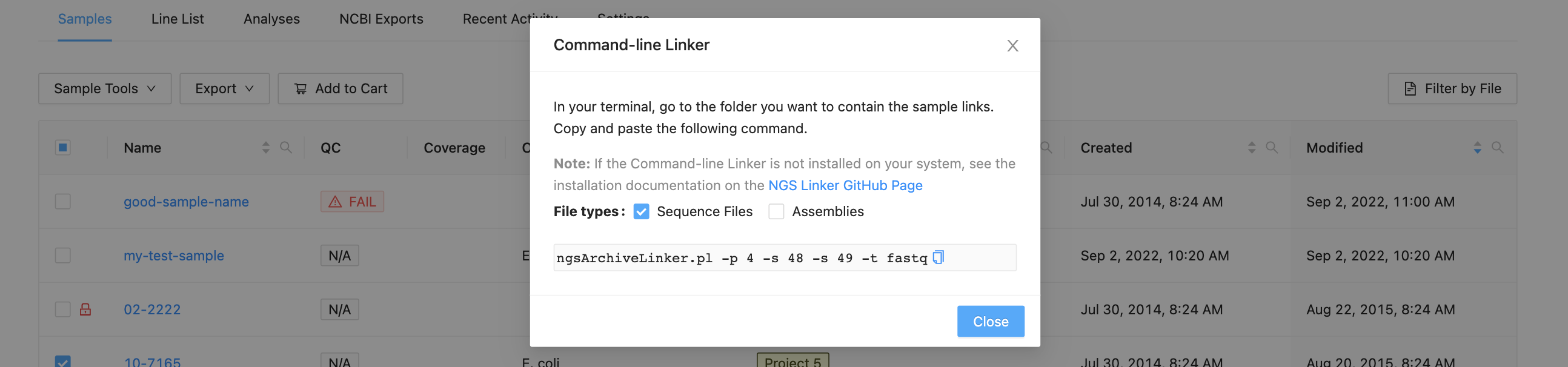
Copy and paste the command into a terminal window and use the username and password that you use to log in to IRIDA:
[user@waffles ~]$ ngsArchiveLinker.pl -p 2 -s 5
Writing files to /home/user
Enter username: user
Enter password:
Reading samples 5 from project 2
Created 2 files for 1 samples in /home/user/Project
[user@waffles ~]$
The folder structure that will be created in the current working directory will match the structure present in IRIDA:
[user@waffles ~]$ tree Project/
Project/
└── sample-1
├── sample-1_S1_L001_R1_001.fastq -> /opt/irida/sequence-files/1/sample-1_S1_L001_R1_001.fastq
└── sample-1_S1_L001_R2_001.fastq -> /opt/irida/sequence-files/2/sample-2_S1_L001_R2_001.fastq
1 directory, 2 files
Importantly, the files that are stored in your directory structure are links and not copies of the files. The purpose of links is to reduce the use of disk space on shared resources. An unfortunate side effect of the link structure is that you cannot change the contents of the files.
