Multi-locus Sequence Typing with MentaLiST
This is a quick tutorial on how to use IRIDA to analyze data with MentaLiST.
- Prepare Kmer Database
- Initial Data
- Adding Samples to the Cart
- Selecting a Pipeline
- Selecting Parameters
- Viewing the Results
- Interpreting the Results
- Viewing Provenance Information
- Viewing Pipeline Details
Prepare Kmer Database
Before analyzing samples, we must prepare a MentaLiST kmer database for the organism of interest. This is done in the Galaxy web interface, and must be done with an account that has Galaxy Admin privileges. This step will only need to be done once per organism, then subsequent analyses can re-use the same kmer database. Please refer to the administrator documentation for detailed instructions on installing MentaLiST kmer databases.
Initial Data
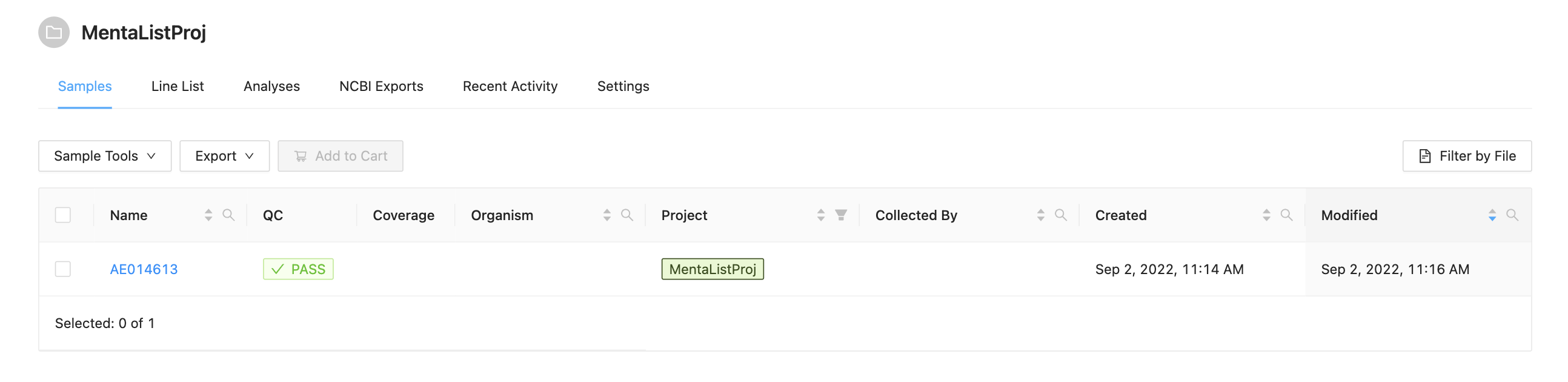
The data for this tutorial comes from https://sairidapublic.blob.core.windows.net/downloads/data/irida-sample-data.zip. It is assumed the sequence files in miseq-run-salmonella/ have been uploaded into appropriate samples as described in the Web Upload Tutorial. Before starting this tutorial you should have a project with samples that appear as:
Adding Samples to the Cart
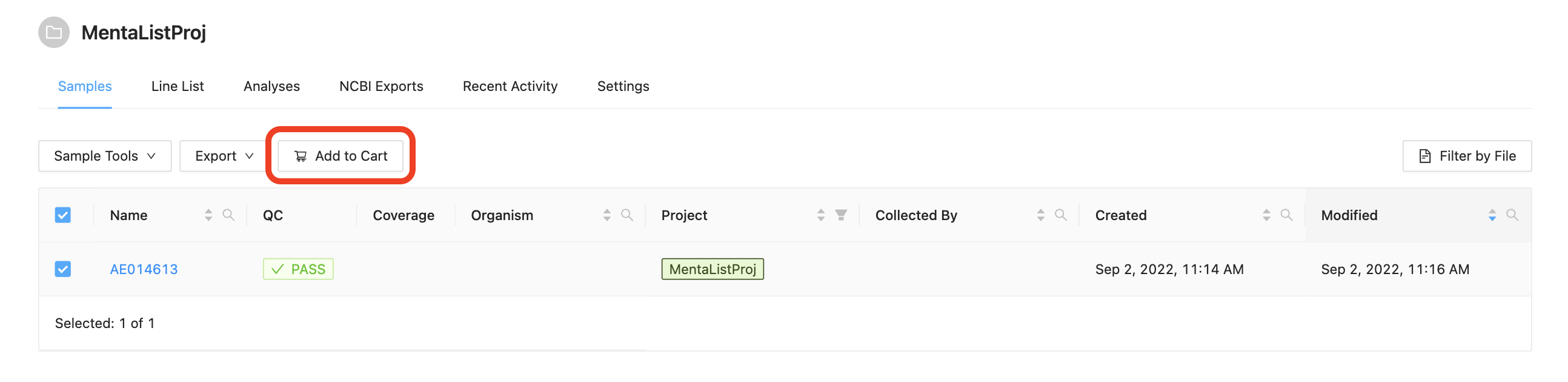
Before a pipeline can be run a set of samples and sequence read data must be selected and added to the cart. For this tutorial please select the single sample and click the Add to Cart button.
Once the samples have been added to the cart, the samples can be reviewed by clicking on the Cart button at the top.
Selecting a Pipeline
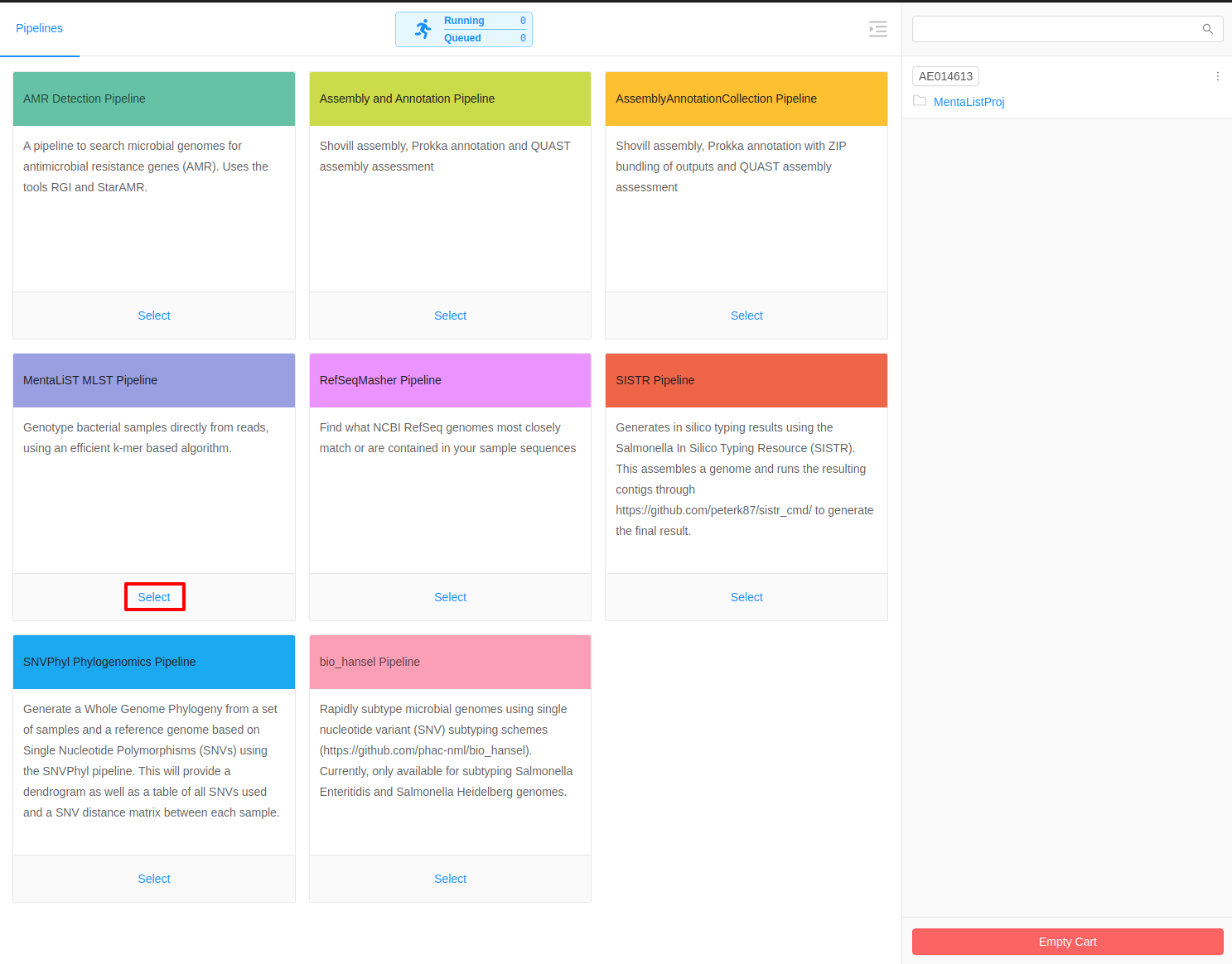
Once inside the cart all available pipelines will be listed in the main area of the page. For this tutorial, we will select the MentaLiST MLST Pipeline.
Selecting Parameters
Once the pipeline is selected, the next page provides an overview of all the input files, as well as the option to modify parameters.
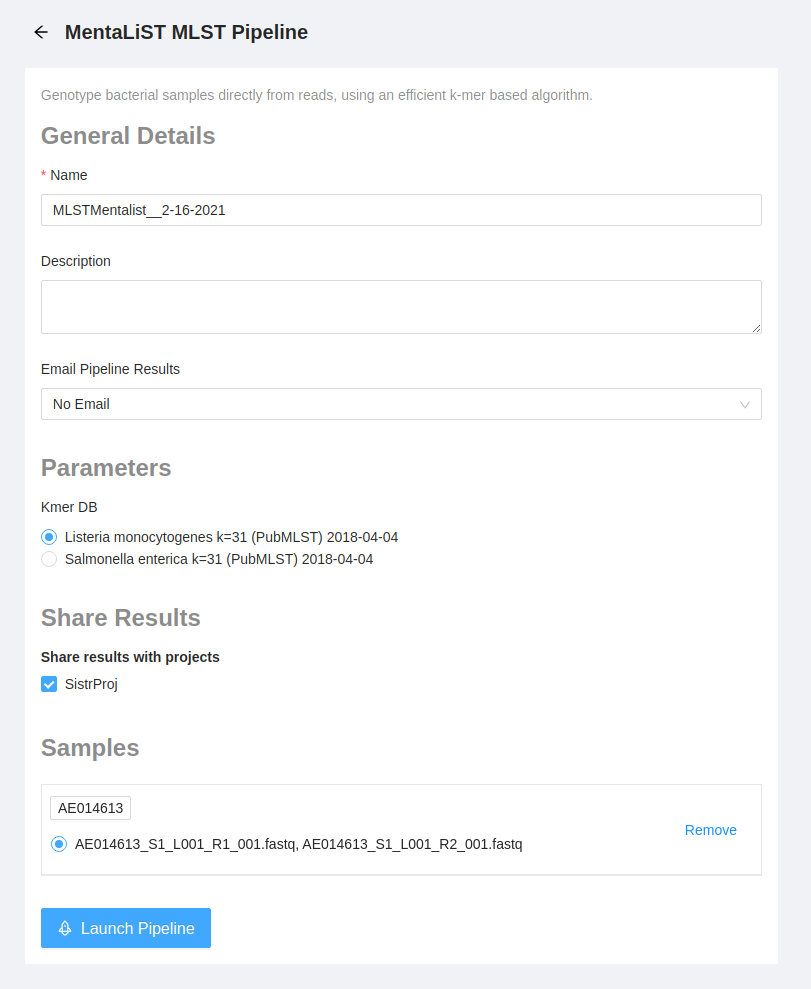
Before launching the pipeline, we must select a MentaLiST kmer database to run our samples against. Select an appropriate ‘Salmonella enterica’ database from the Kmer DB from the options displayed. Note that the kmer databases available on your system will have a different date and may have been built with a different value of k than the ones shown below. If there is no ‘Salmonella enterica’ database available, please contact your IRIDA system administrator and refer to the administrator documentation for detailed instructions on installing MentaLiST kmer databases.

Please use the Launch Pipeline button to start the pipeline.

Once the button is selected, you will be redirected to the analysis details page.
Viewing the Results
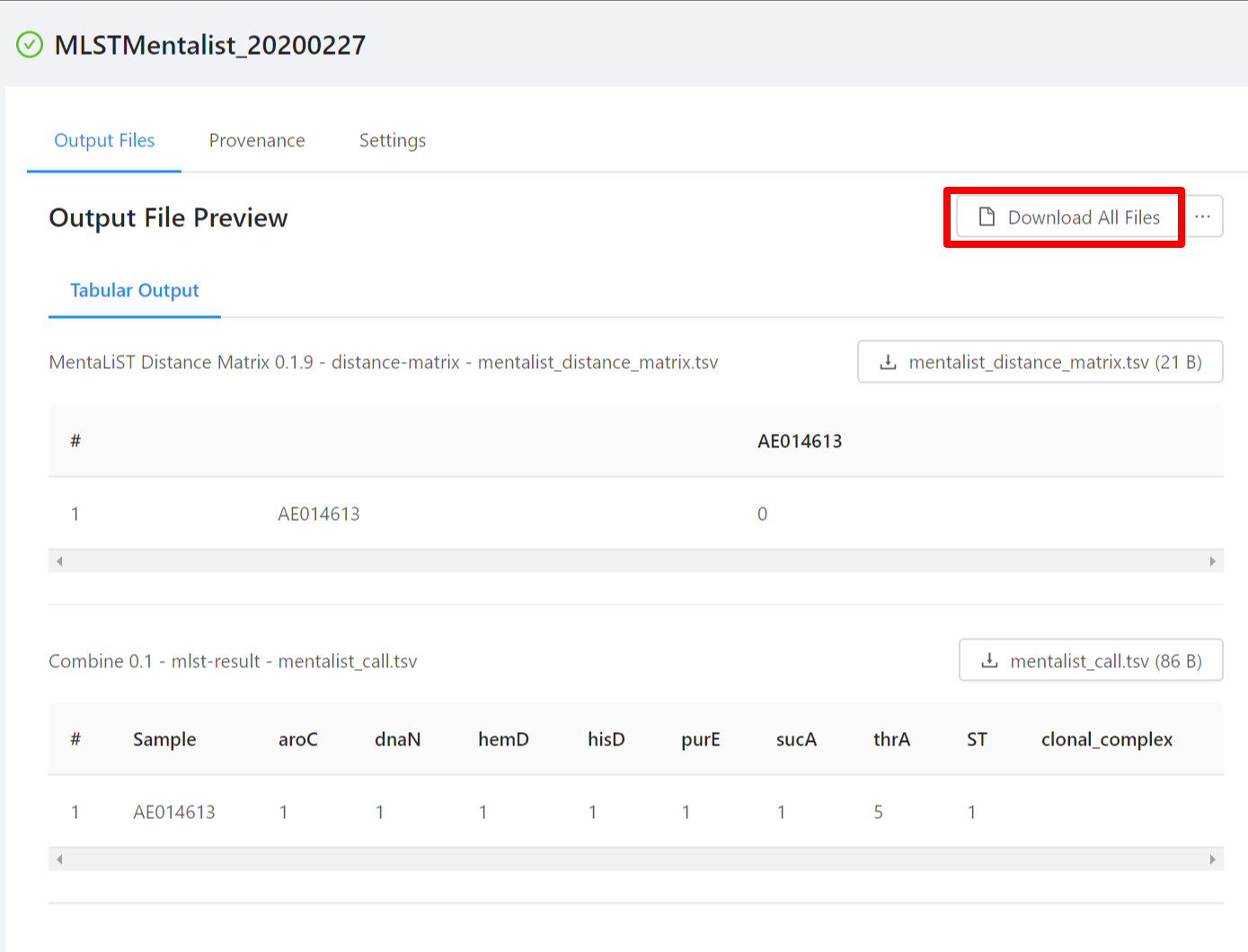
Once the pipeline is complete, you will be able to view the results of the MentaList analysis. Note that not all files have an available preview and as such are not displayed in the Output File Preview but are downloaded when selecting the Download All Files button.
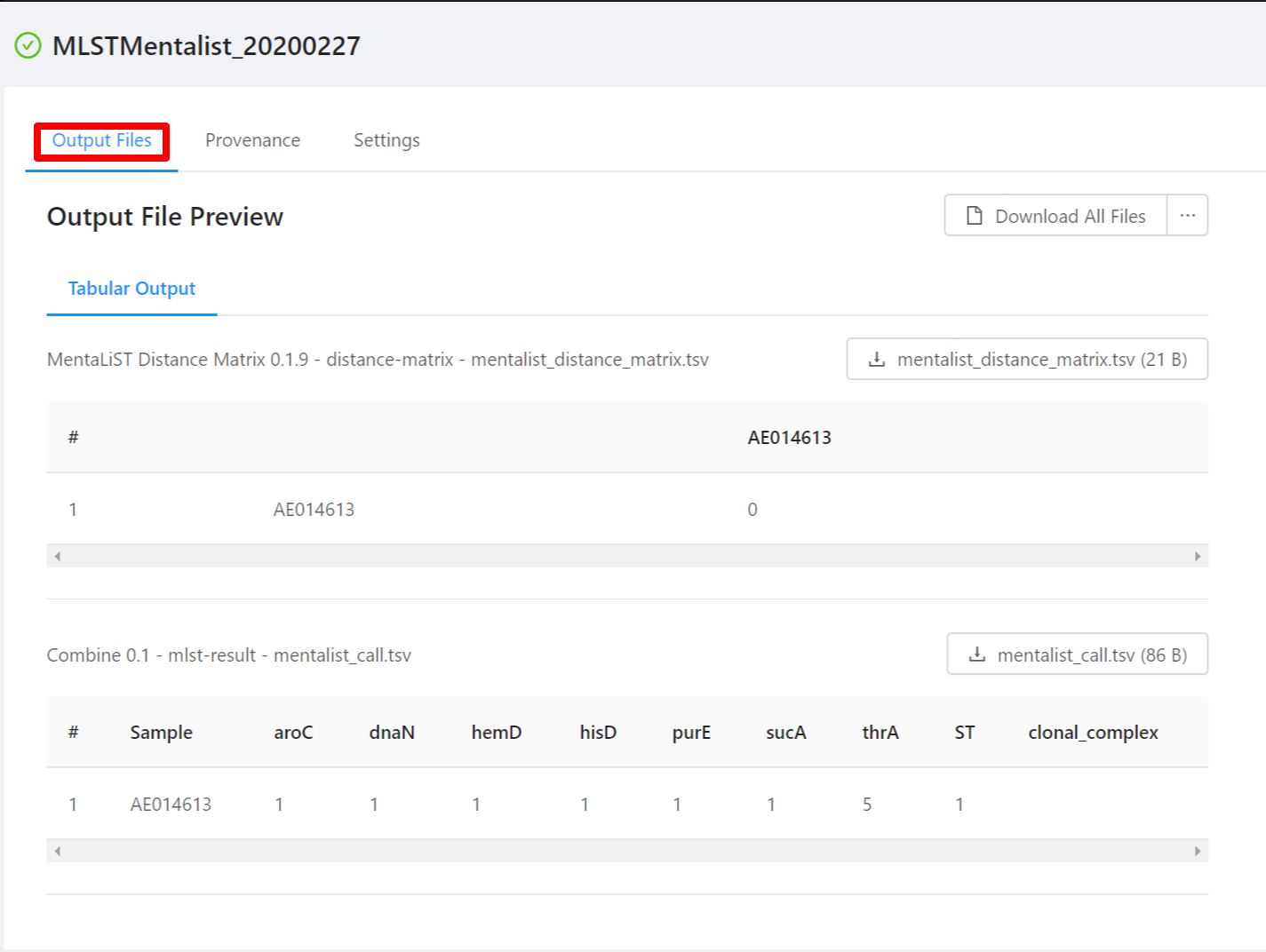
To download individual files select the … next to the Download All Files and select the file to download.
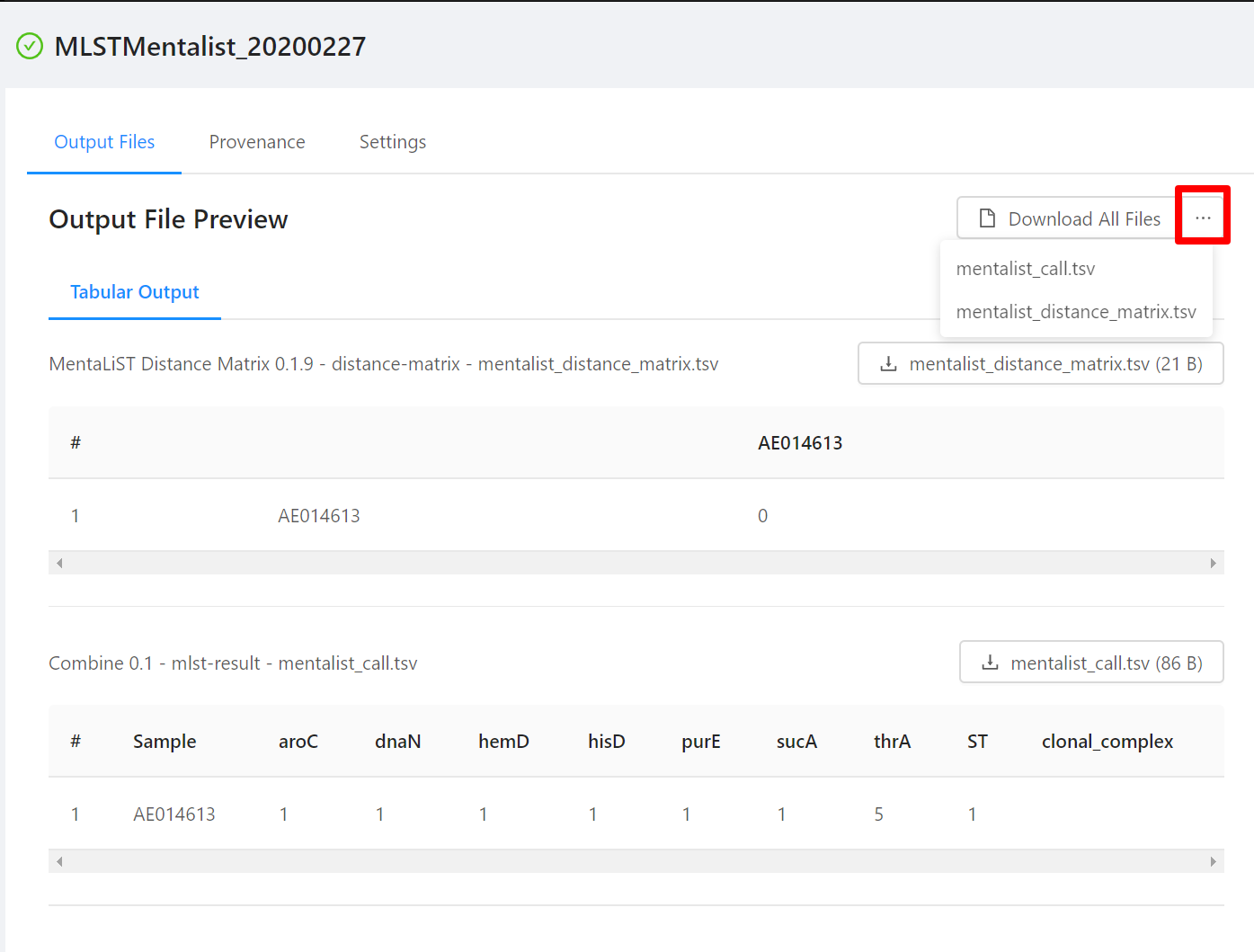
To download al the files generated by the analysis, please select the Download All Files button.
Interpreting the Results
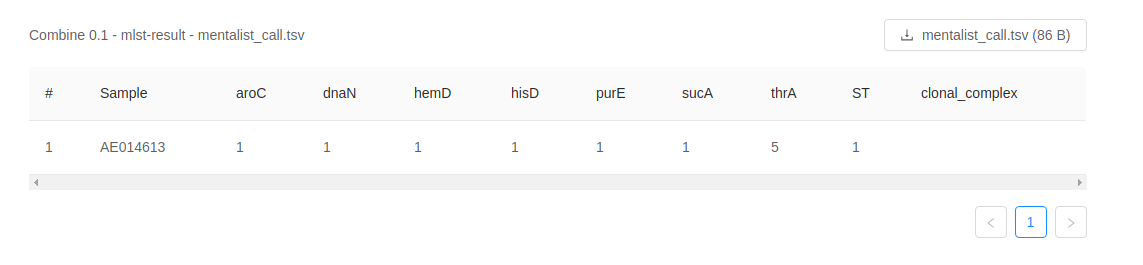
MentaLiST provides MLST results in a simple tab-separated value file that can be opened in a plaintext editor such as Microsoft Notepad or Apple TextEdit, or a spreadsheet application such as Microsoft Excel or LibreOffice Calc.
The first row is a header. The following rows represent the MLST results for each sample. The first column contains the sample ID. Subsequent columns contain allele calls for each locus in the typing scheme. The sequence type (ST) and clonal complex are reported in the final two columns.
Viewing Provenance Information
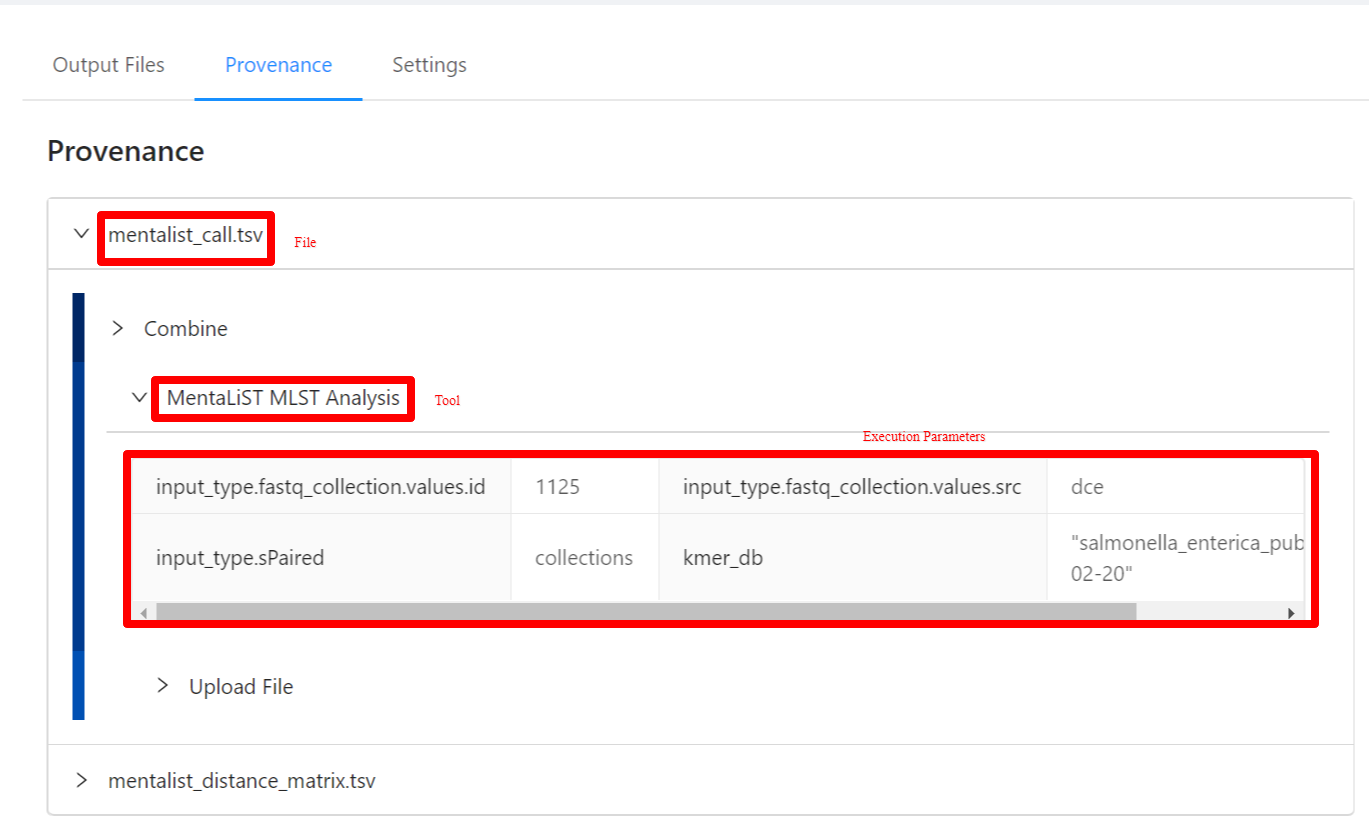
To view the pipeline provenance information, please select the Provenance tab.
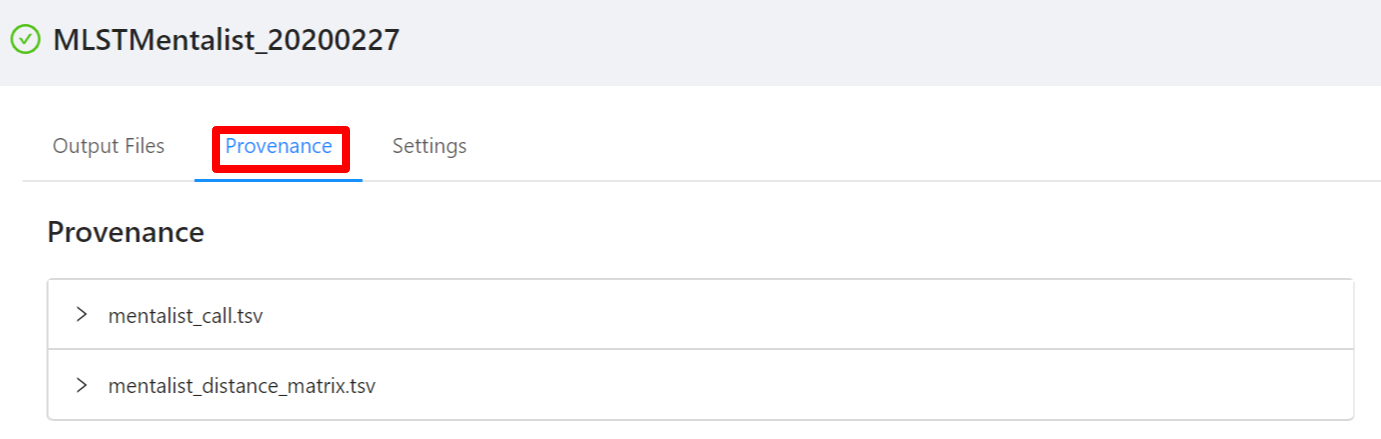
The provenance is displayed on a per file basis. Clicking on the mentalist_call.tsv file will display it’s provenance. Expanding each tool will display the parameters that the tool was executed with.
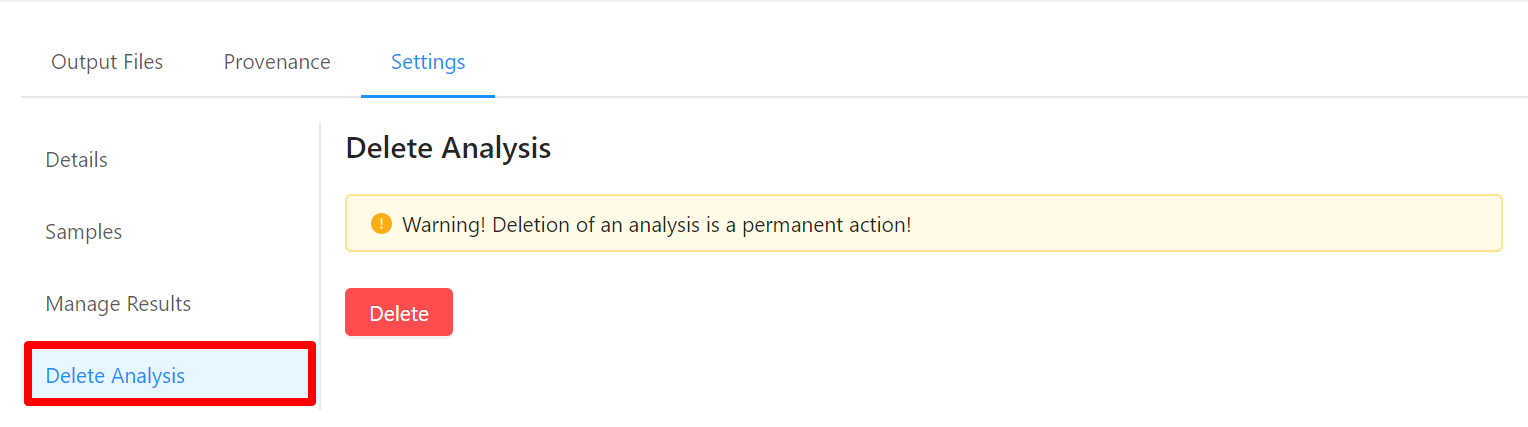
Viewing Pipeline Details
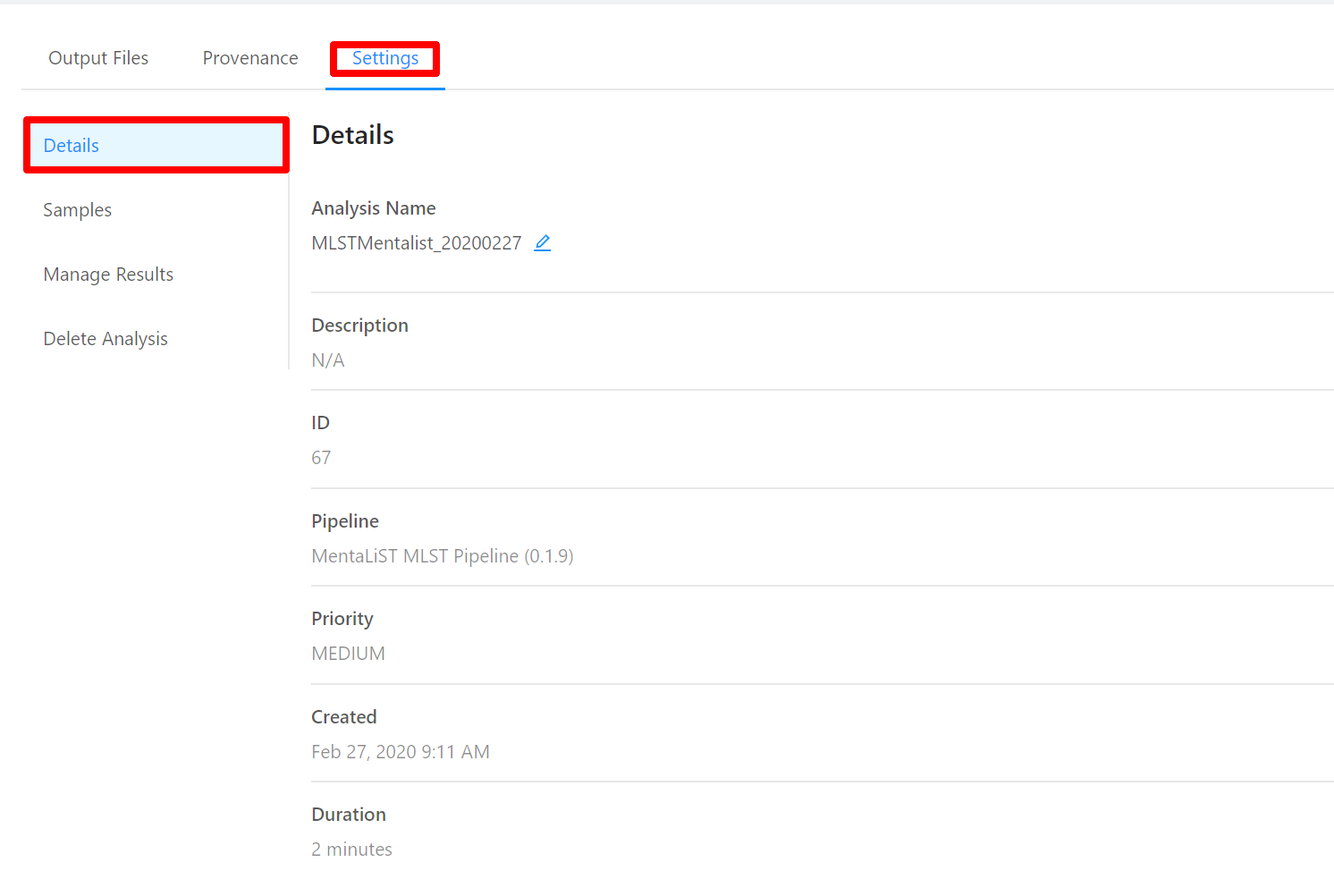
To view analysis details, please select the Settings tab. From here you can view the analysis name, analysis description, analysis id, pipeline and pipeline version used by the analysis, analysis priority, when the analysis was created, and duration of the analysis.
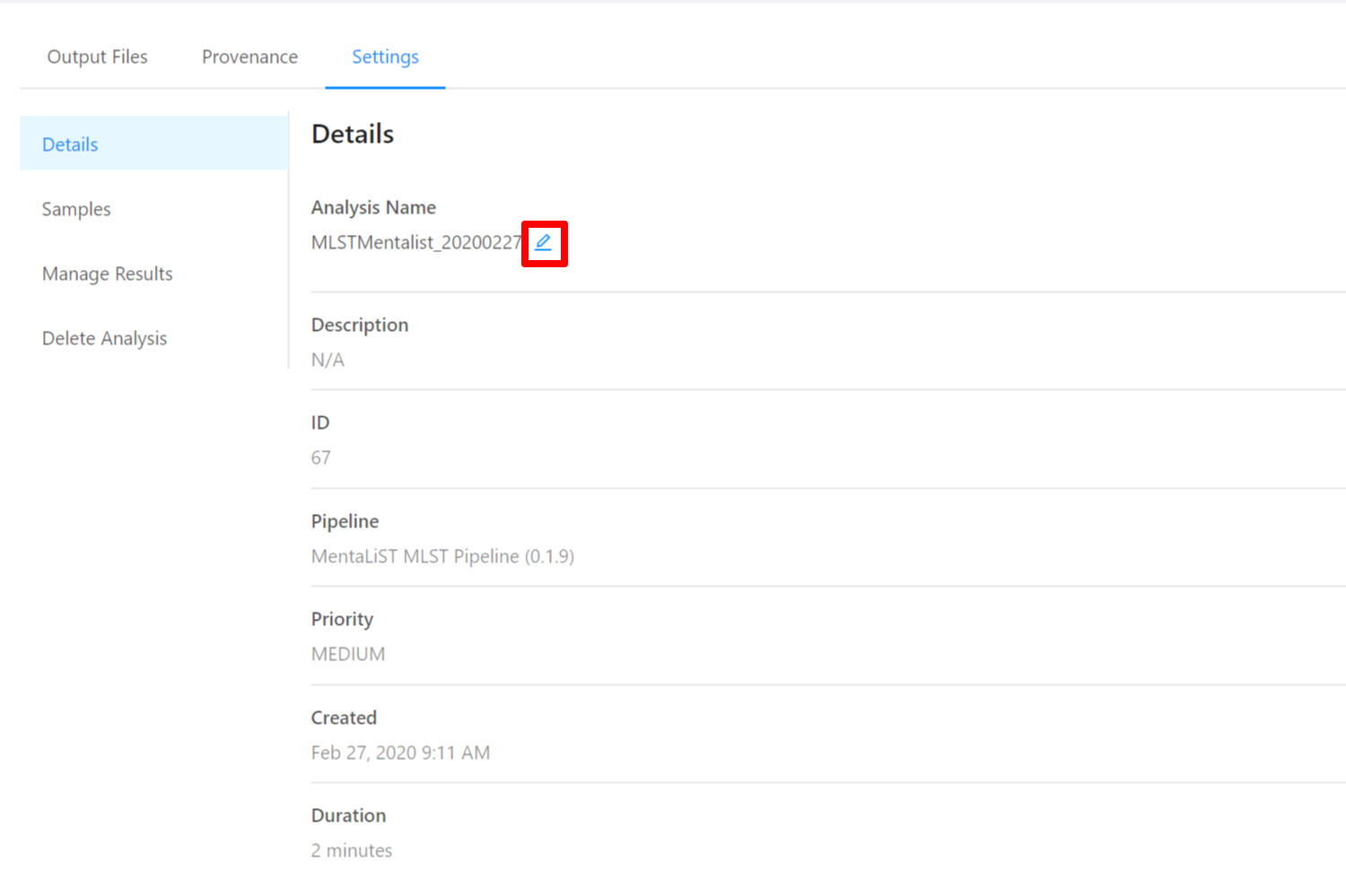
To edit an analysis name, please select the Pencil icon next to the analysis name. Once you have edited the analysis name, pressing the ENTER key on your keyboard or clicking anywhere outside of the text box will update the name. To cancel editing a name you can either hit the ESC key on your keyboard or if the name has not been changed you can also click anywhere outside of the text box.
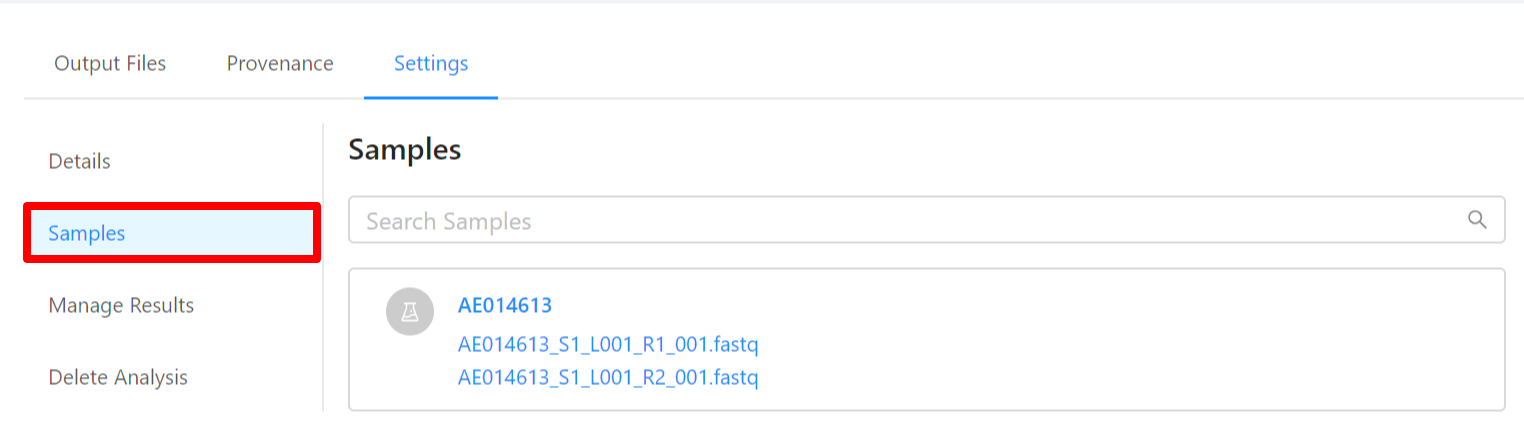
To view samples used by the analysis, please select the Samples tab.
To share analysis results with other projects, please select the Manage Results tab.
To delete an analysis, please select the Delete Analysis tab.